We propose SASNet, a dual-branch semi-supervised segmentation network that adaptively fuses multi-scale features to improve performance under limited annotation. • A scale-aware adaptive reweight strategy is introduced to generate more reliable ensemble predictions by selectively fusing pixel-wise results. • A view variance enhancement mechanism simulates annotation differences across views and scales, improving robustness and segmentation accuracy.
In medical image segmentation, semi-supervised learning is crucial because high-quality dense annotations are both expensive and limited. At the same time, due to differences in the level of annotations, there may be some domain offset between them, resulting in less label information that can be used [1]. Therefore, more and more researchers
Low-level branch Dual-branch have begun to combine semi-supervised learning with medical image segmentation in recent years. Among them, Chaitanya et al. [2] designed a local contrast loss to help the model learn target features and generate better pseudo labels. In addition, many researchers have also begun to study the introduction of regularization terms into the loss function to help improve the performance of semi-supervised learning. Luo et al. [3] constructed a joint framework that utilizes CNN and Transformer structure to learn different features of images and uses their prediction results for mutual supervision. You et al. [4] used the teacher network and student network to calculate the comparative loss between the predicted results of the two networks. At the same time, researchers are also attempting to utilize multi-scale learning to improve the performance of medical image segmentation. Liu et al. [5] directly input data of different scales into the encoder and output prediction results of different scales in the decoder section. Wang et al. [6] developed a multi-scale fusion module, which helps to fuse spatial information of different scales through route convolution. But researchers often ignore the role of multi-scale learning in semi-supervised learning [7][8][9], so we design a new multi-scale learning paradigm-Scale-aware Adaptive Learning and innovatively introduce it into semi-supervised learning.
As we mentioned above, there are still many unresolved issues in semi-supervised medical image segmentation.
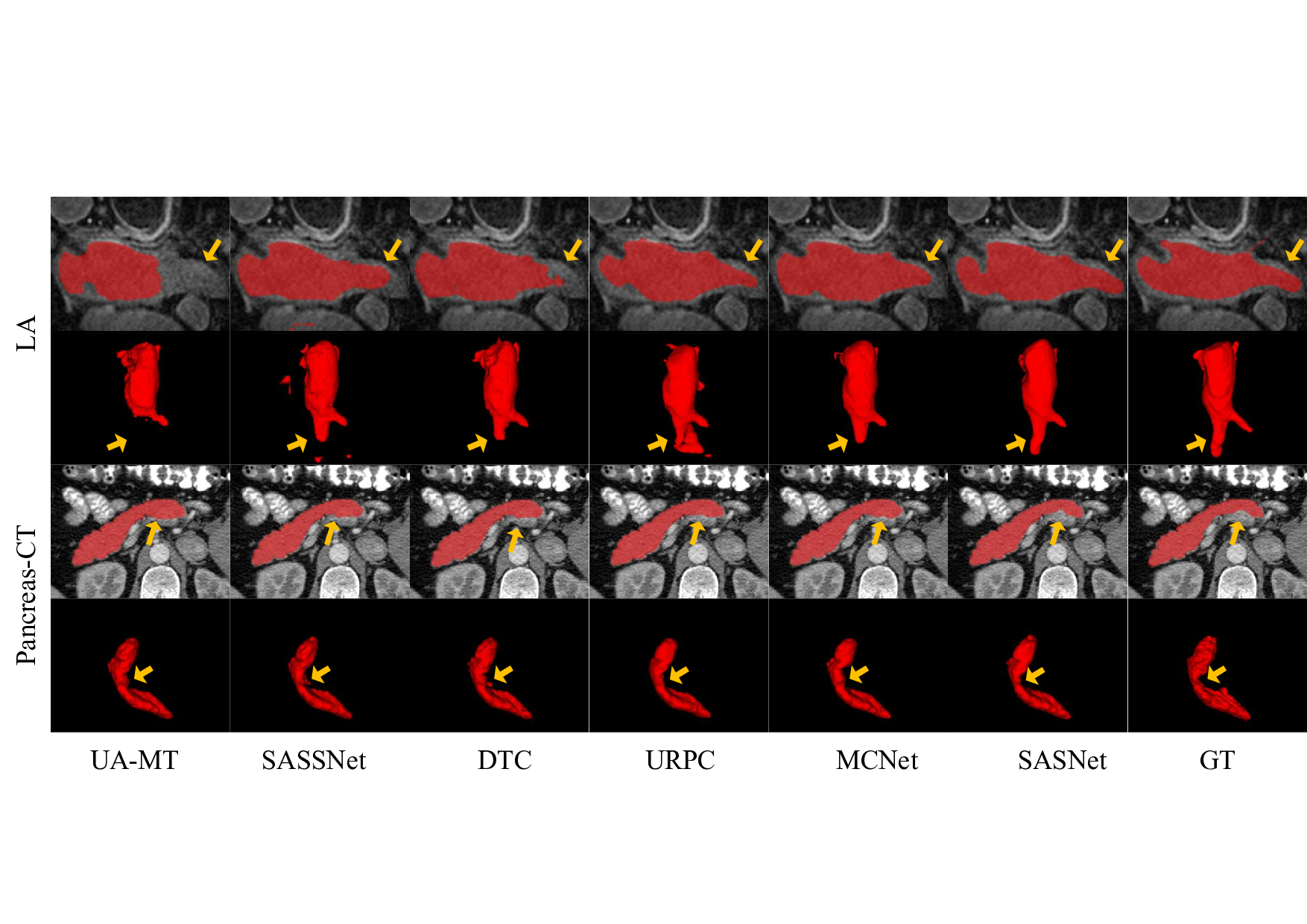
Firstly, the challenge of missing labels remains a significant issue, as semi-supervised methods continue to demonstrate lower performance in comparison to fully supervised approaches. Specifically, the segmentation of small targets and boundaries remains suboptimal, so we decide to further explore multi-scale information in the data to compensate for the missing labels. Incorporating feature information at multiple scales can enhance the model’s ability to identify small targets and refine segmentation boundaries. Secondly, the problem of annotation variance poses a unique challenge in the field of medical imaging, given the varying focus. The difference in annotations can exacerbate the challenge of limited labeled data, especially when we need to refine predictions of boundaries.
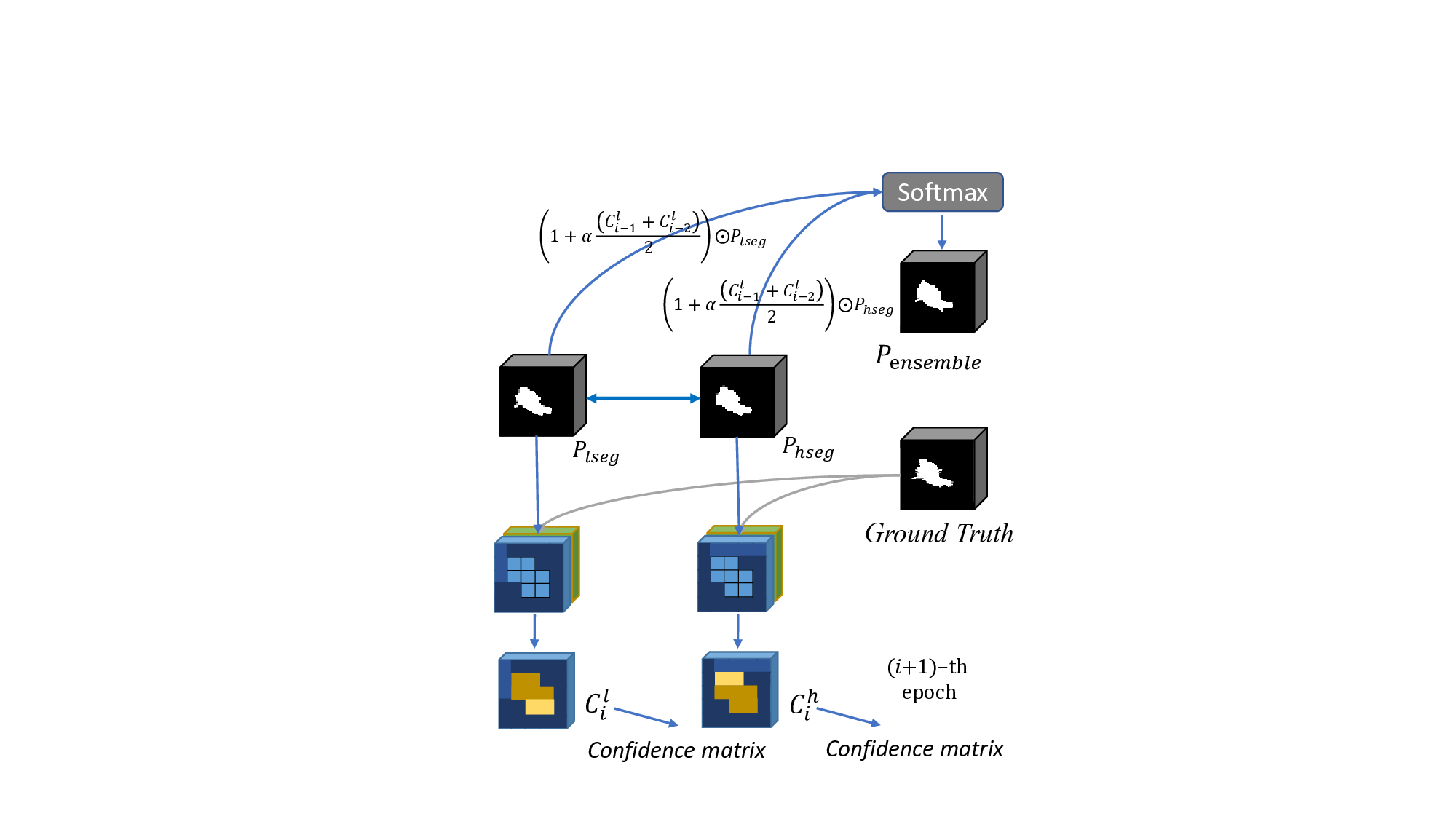
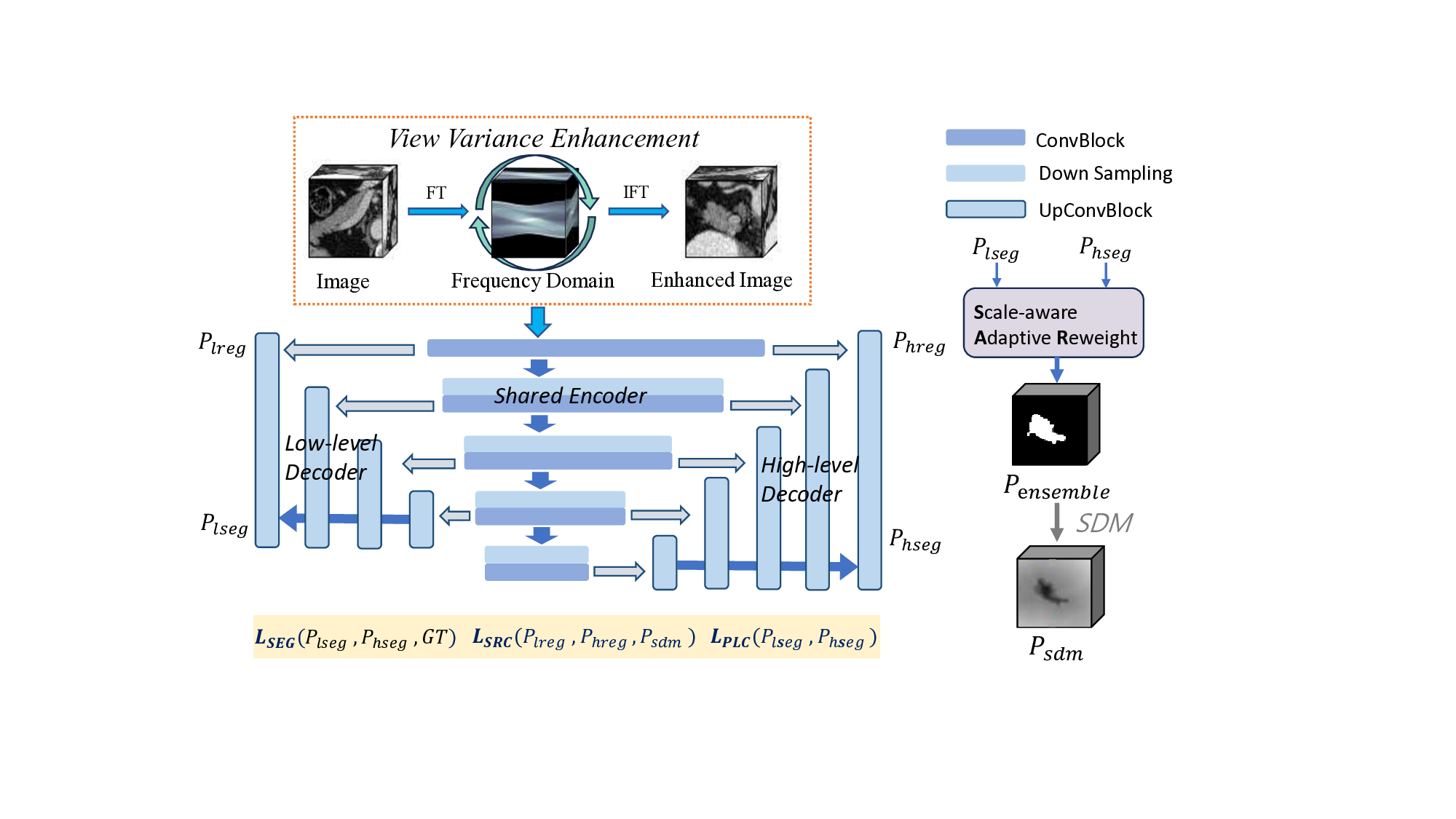
To address the above issues, we propose an Adaptive Supervised Hierarchical Network based on Scale Invariance. Specifically, to enhance the model’s proficiency in learning multi-scale information, we design two different branches, one focusing on low-level features and the other on high-level features. Unlike previous multi-scale learning networks, we do not use multi-scale data input to different encoders but instead utilize multi-scale encoding features input to different decoders. We believe that it can obtain high-quality encoding features. In addition, directly utilizing encoding features at different scales can more intuitively utilize the information at different scales and display their common areas of interest as shown in Fig. 1, which we call it Scale Invariance. The figure illustrates that both low-level and high-level features are utilized separately, ignoring the predicted connectivity information. Using them simultaneously can achieve the precise localization of the target, and enrich the model with more semantic information. As demonstrated in Fig. 1, our model exhibits distinct prediction styles across different views, akin to the natural variability among predictions. It benefits from limited labeled data, aiming to enhance the model’s robustness by incorporating view variance. To address the above challenges, a natural consideration arises-integrating the results from both branches to achieve a more comprehensive and accurate segmentation. However, a simple addition of the two results may introduce undesirable noise, potentially compromising overall performance. Therefore, we introduce the Scale-aware Adaptive Reweight (SAR) strategy. This approach involves pixel-wise weighting of predictions from both branches during the training process, based on the confidence acquired from preceding epochs.
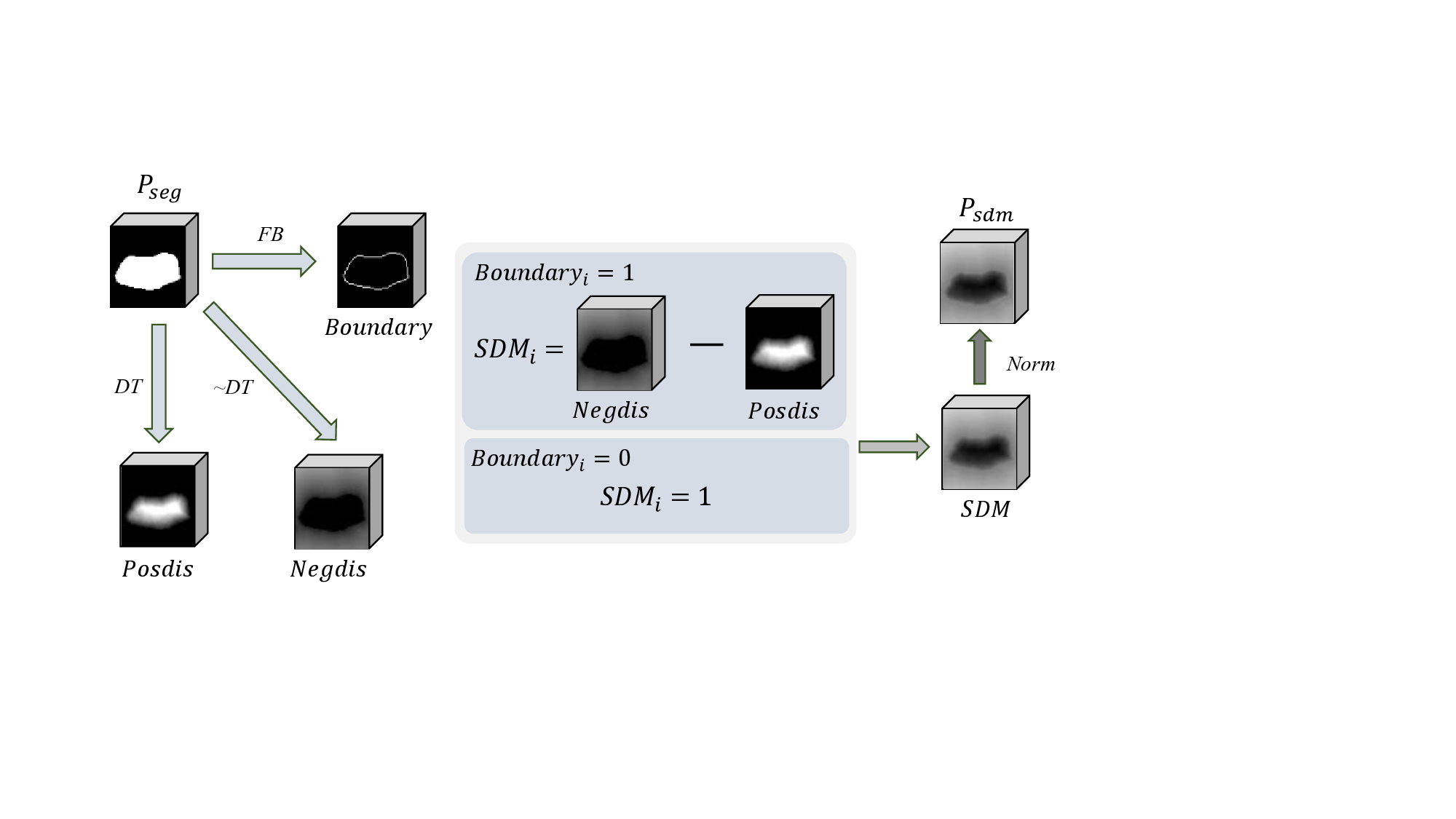
This adaptive mechanism empowers the network to selectively favor more reliable results, mitigating potential errors introduced through direct addition. The incorporation of the SAR strategy refines and controls the fusion process, resulting in a notable enhancement in overall segmentation performance. To address the challenge of annotation variance, we introduce a view variance enhancement approach to simulate annotation differences. As shown in Fig. 1, b
This content is AI-processed based on open access ArXiv data.