We present PathoSyn, a unified generative framework for Magnetic Resonance Imaging (MRI) image synthesis that reformulates imaging-pathology as a disentangled additive deviation on a stable anatomical manifold. Current generative models typically operate in the global pixel domain or rely on binary masks, these paradigms often suffer from feature entanglement, leading to corrupted anatomical substrates or structural discontinuities. PathoSyn addresses these limitations by decomposing the synthesis task into deterministic anatomical reconstruction and stochastic deviation modeling. Central to our framework is a Deviation-Space Diffusion Model designed to learn the conditional distribution of pathological residuals, thereby capturing localized intensity variations while preserving global structural integrity by construction. To ensure spatial coherence, the diffusion process is coupled with a seam-aware fusion strategy and an inference-time stabilization module, which collectively suppress boundary artifacts and produce high-fidelity internal lesion heterogeneity. PathoSyn provides a mathematically principled pipeline for generating high-fidelity patient-specific synthetic datasets, facilitating the development of robust diagnostic algorithms in low-data regimes. By allowing interpretable counterfactual disease progression modeling, the framework supports precision intervention planning and provides a controlled environment for benchmarking clinical decision-support systems. Quantitative and qualitative evaluations on tumor imaging benchmarks demonstrate that PathoSyn significantly outperforms holistic diffusion and mask-conditioned baselines in both perceptual realism and anatomical fidelity. The source code of this work will be made publicly available.
support [7], [8]. However, acquiring balanced datasets is hindered by limited patient availability, ethical constraints, and high annotation costs. Generative models mitigate this scarcity by synthesizing realistic pathological images that complement real-world data. Beyond algorithmic utility, highfidelity synthetic pathology aids in clinical intervention planning by illustrating plausible disease trajectories, surfacing rare edge cases, and enabling controlled evaluation of models prior to deployment [4], [5], [9]- [11]. Enhancing the fidelity and consistency of these images is therefore directly linked to developing safer, more reliable tools for clinical decisionmaking.
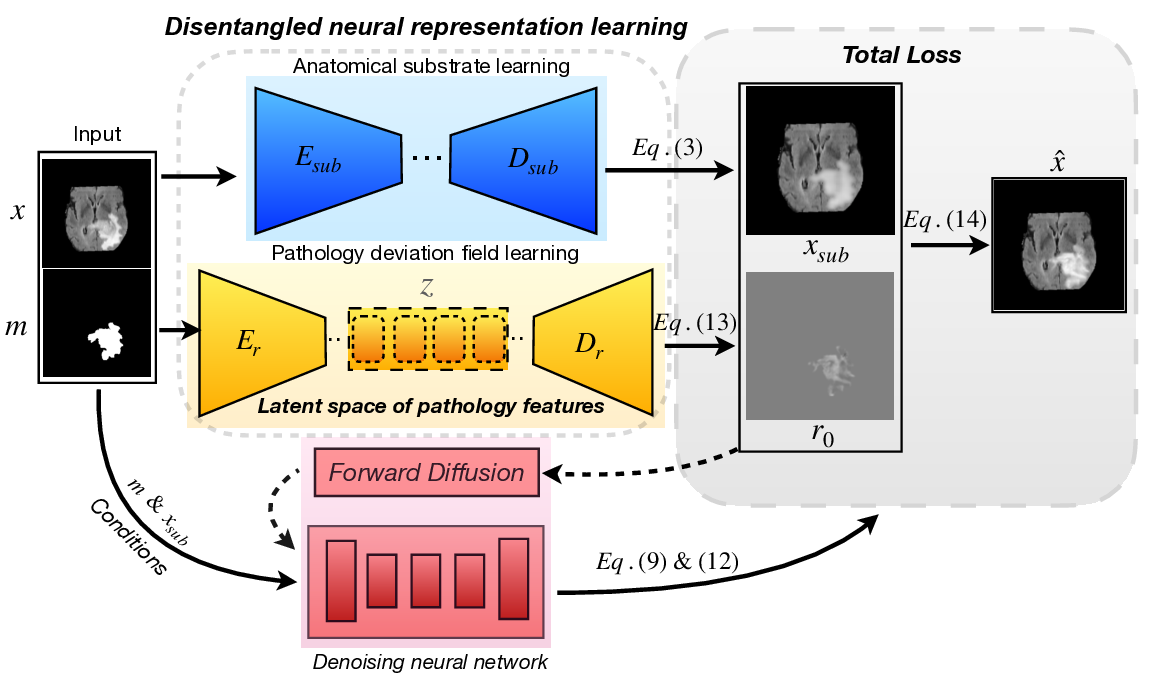
Despite growing interest, generating pathological images remains challenging due to the intrinsic asymmetry between anatomy and disease. The anatomical structure, including organ geometry and spatial layout, is largely stable for a given subject, whereas the pathological appearance is the main source of uncertainty, varying in intensity, texture, and clinical stage. The most clinically relevant variability is concentrated in the way disease perturbs a stable anatomical substrate. Existing generative models [3], [12] rarely make this distinction explicit. When operating directly in image space, anatomy and pathology are treated equally stochastic; the model must learn a high-dimensional distribution over all pixels simultaneously. This unnecessarily enlarges the solution space and permits the generator to introduce anatomical modifications in order to fit the data, thereby producing systematic distortions of anatomically stable structures. Consequently, current approaches fall into two suboptimal extremes: either they model the image as a holistic entity with no explicit separation [13], or they enforce complete separation via masking, which identifies location, but overlooks internal lesion appearance and continuity with surrounding tissue.
Although deep learning-based methods have significantly advanced visual fidelity compared to traditional rule-based algorithms, they have remained largely confined to the same image-space paradigm [14], [15]. Early applications of generative adversarial networks (GANs) synthesized lesions by modifying entire slices or volumes, sometimes conditioned on labels or coarse masks [1], [16]- [19]; these models can create sharp and visually plausible images, but tend to be unstable, difficult to control, and prone to hallucination of anatomy that does not correspond to any realistic background. Variational autoencoders (VAEs) introduced a probabilistic latent representation and more stable training [20]- [22], and structured or disentangled variants attempted to separate anatomy and pathology in latent space [23], [24]. However, because decoding still happens directly to full images, they often oversmooth fine pathological details and leak stochastic variation into non-lesional regions. More recently, diffusion models have become the dominant paradigm for high-fidelity synthesis in both natural and medical imaging [12], [13], [25]- [27]. By iteratively denoising from a simple prior, they capture rich, multimodal image statistics and offer strong sample diversity. However, most medical diffusion models still learn a distribution over entire images or over mask-conditioned images, without explicitly isolating where uncertainty should reside. Mask conditioning helps to specify where pathology should appear but not what it should look like internally [28]- [30]; lesions may have plausible boundaries, but lack realistic internal texture, heterogeneity, or progression patterns. As a result, even advanced models still entangle anatomical background with pathological variation and fail to exploit that anatomy is largely deterministic while pathology is a structured deviation (Fig. 1).
These limitations indicate that progress in pathological synthesis is not primarily determined by the choice of network architecture but by how the problem itself is represented. When a model treats the entire image as an unconstrained random variable, it must learn a full high-dimensional distribution in which anatomy and pathology vary simultaneously, even though only one of them is expected to change. Instead, a more principled formulation views a pathological image as the combination of two qualitatively different factors: a stable anatomical substrate that should remain preserved and a pathological deviation that carries the uncertainty, diversity and progression of the disease. Structurally separating these components reduces the effective complexity of the generative task and better reflects clinical reality: anatomy behaves as the baseline, while pathology introduces variation. In other words, disease does not replace the entire image; it perturbs an existing anatomical state. Motivated by this perspective, we introduce PathoSyn, a deviation-based framework for pathological image generation. Rather than end-to-end synthesizin
This content is AI-processed based on open access ArXiv data.