There has been unprecedented interest in developing agents that expand the boundary of scientific discovery, primarily by optimizing quantitative objective functions specified by scientists. However, for grand challenges in science , these objectives are only imperfect proxies. We argue that automating objective function design is a central, yet unmet requirement for scientific discovery agents. In this work, we introduce the Scientific Autonomous Goal-evolving Agent (SAGA) to amend this challenge. SAGA employs a bi-level architecture in which an outer loop of LLM agents analyzes optimization outcomes, proposes new objectives, and converts them into computable scoring functions, while an inner loop performs solution optimization under the current objectives. This bi-level design enables systematic exploration of the space of objectives and their trade-offs, rather than treating them as fixed inputs. We demonstrate the framework through a broad spectrum of applications, including antibiotic design, inorganic materials design, functional DNA sequence design, and chemical process design, showing that automating objective formulation can substantially improve the effectiveness of scientific discovery agents.
Scientific discovery has been driven by human ingenuity through iterations of hypothesis, experimentation, and observation, but is increasingly bottlenecked by the vast space of potential solutions to explore and the high cost of experimental validation. Recent advances in artificial intelligence (AI) agents based on large language models (LLMs) offer promising approaches to address these bottlenecks and accelerate scientific discovery. Leveraging massive pretrained knowledge and general capabilities for information collection and reasoning, these AI agents can efficiently navigate large solution spaces and reduce experimental costs by automating key aspects of the research process. For example, pipeline automation agents [ specialized data analysis workflows, reducing the manual effort required for routine experimental processes. AI Scientist agents [3,4,5,6,7] tackle the exploration challenge by autonomously generating and evaluating novel hypotheses (e.g., the relationship between a certain mutation and a certain disease) through integrated literature search, data analysis, and academic writing capabilities.
Our work embarks on a different and more ambitious goal in scientific discovery: building agents to discover new solutions to complex scientific challenges, such as proofs for conjectures, faster algorithms, better therapeutic molecules, and functional materials. This problem is uniquely challenging due to the “creativity” and “novelty” required and the infinite combinatorial search space for potential solutions. Previous work has sought to address these challenges by developing optimization models that automatically find solutions maximizing a manually defined set of quantitative objectives, such as drug efficacy, protein expression, and material stability. These approaches, ranging from traditional generative models to more recent LLM-based methods, have demonstrated the ability to efficiently optimize against fixed objectives in domains including drug design [8], algorithm discovery [9], and materials design [10].
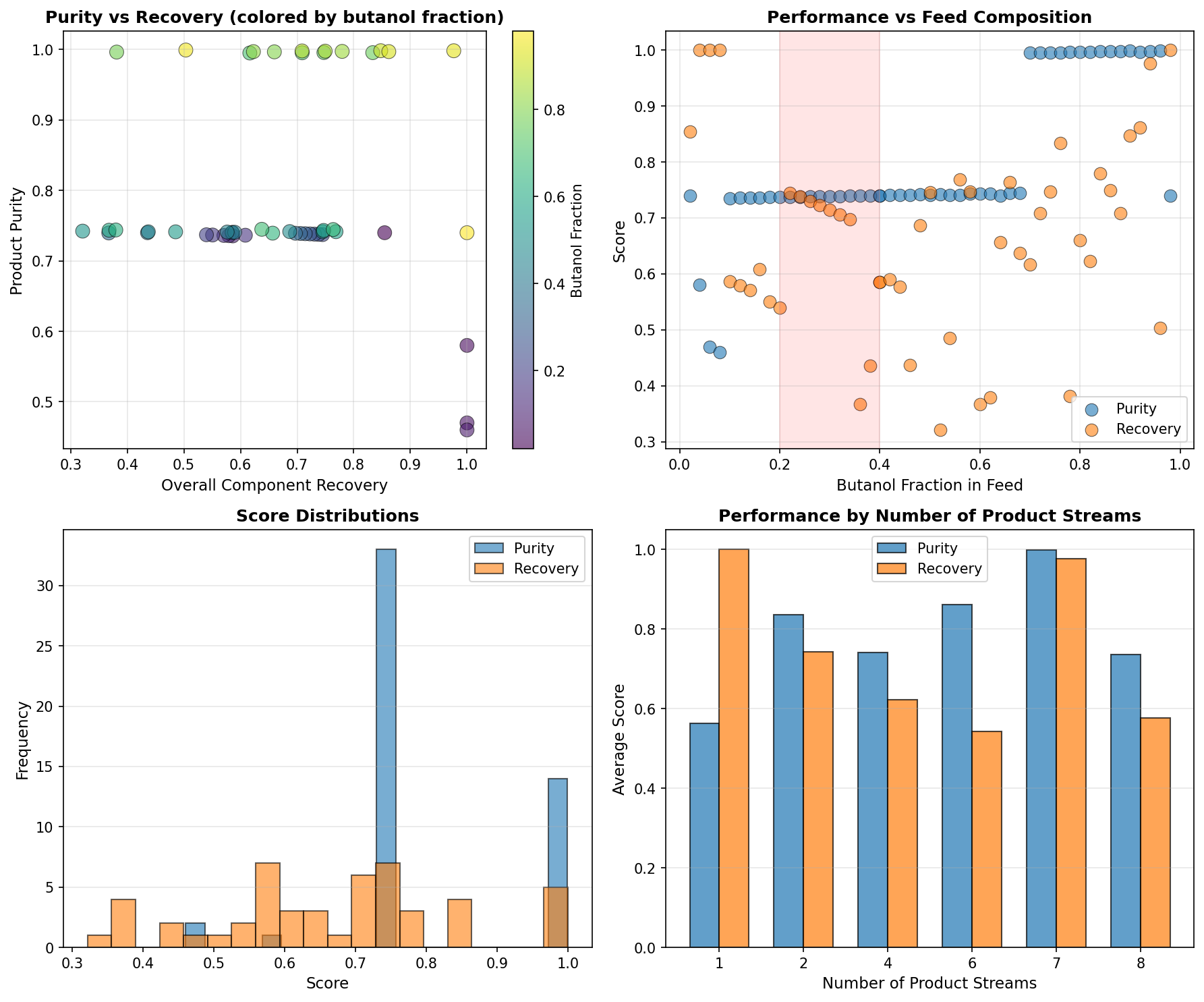
However, these optimization models operate under a critical assumption: that the right set of objective functions is known upfront. In practice, this assumption rarely holds. Just as scientific discovery requires iterations of hypothesis, experimentation, and observation, determining the appropriate objectives for a discovery task is itself an iterative search process. Scientists must constantly tweak objectives based on intermediate results, domain knowledge, and practical constraints that emerge during exploration (Figure 1(a)). This iterative refinement is particularly crucial in experimental disciplines such as drug discovery, materials design, and protein engineering, where many critical properties can only be approximated through predictive models. Without this evolving process, the discovery suffers from reward hacking issues [11]: they exploit gaps between models and reality, producing solutions that maximize predicted scores while missing important practical considerations that experts would recognize. The search space for objectives and their relative weights is itself combinatorially large (Figure 1(b)), making it extremely difficult to specify the right objectives from the outset. As a result, while existing optimization models can solve the low-level optimization problem efficiently, scientific discovery remains bottlenecked by the high-level objective search process that relies on manual trial-and-error.
In this work, we introduce SAGA as our first concrete step toward automating this iterative objective evolving process. SAGA is designed to navigate the combinatorial search space of objectives by integrating high-level objective planning in the outer loop with low-level optimization in the inner loop (Figure 1(c)). The outer loop comprises four agentic modules: a planner that proposes new objectives based on the task goal and current progress, an implementer that converts proposed objectives into executable scoring functions, an optimizer that searches for candidate solutions maximizing the specified objectives, and an analyzer that examines the optimization results and identifies areas for improvement. Within the optimizer module, an inner loop employs any optimization methods (e.g., genetic algorithms or reinforcement learning) to iteratively evolve candidate solutions toward the current objectives. Importantly, SAGA is a flexible framework supporting different levels of human involvement. It offers three modes (Figure 1(d)): (1) co-pilot mode, where scientists collaborate with both the planner and analyzer to reflect on results and determine new objectives; (2) semi-pilot mode, where scientists provide feedback only to the analyzer; and (3) autopilot mode, where both analysis and planning are fully automated. This design allows scientists to interact with SAGA in ways that best suit their expertise and preferences.
SAGA is a generalist scientific discovery agentic f
This content is AI-processed based on open access ArXiv data.