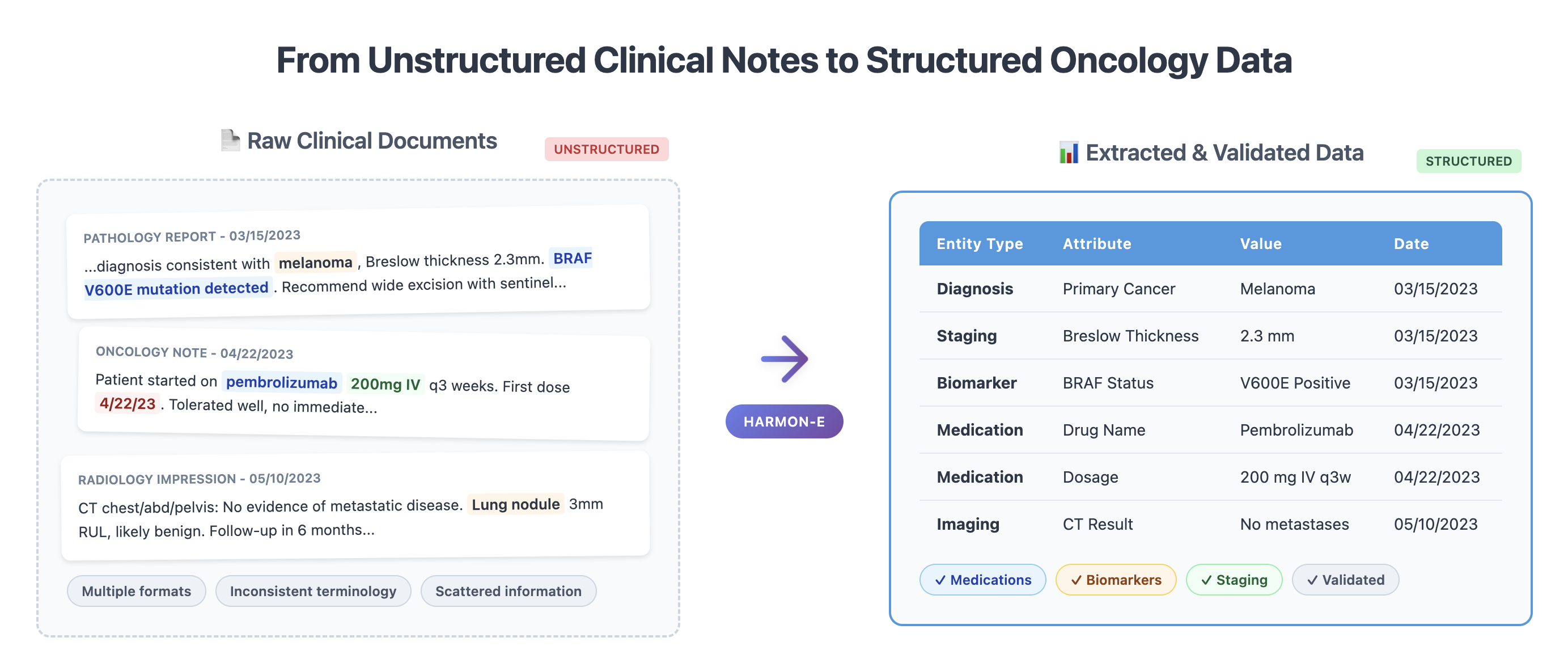
Unstructured notes within the electronic health record (EHR) contain rich clinical information vital for cancer treatment decision making and research, yet reliably extracting structured oncology data remains challenging due to extensive variability, specialized terminology, and inconsistent document formats. Manual abstraction, although accurate, is prohibitively costly and unscalable. Existing automated approaches typically address narrow scenarios - either using synthetic datasets, restricting focus to document-level extraction, or isolating specific clinical variables (e.g., staging, biomarkers, histology) - and do not adequately handle patient-level synthesis across the large number of clinical documents containing contradictory information. In this study, we propose an agentic framework that systematically decomposes complex oncology data extraction into modular, adaptive tasks. Specifically, we use large language models (LLMs) as reasoning agents, equipped with context-sensitive retrieval and iterative synthesis capabilities, to exhaustively and comprehensively extract structured clinical variables from real-world oncology notes. Evaluated on a large-scale dataset of over 400,000 unstructured clinical notes and scanned PDF reports spanning 2,250 cancer patients, our method achieves an average F1-score of 0.93, with 100 out of 103 oncology-specific clinical variables exceeding 0.85, and critical variables (e.g., biomarkers and medications) surpassing 0.95. Moreover, integration of the agentic system into a data curation workflow resulted in 0.94 direct manual approval rate, significantly reducing annotation costs. To our knowledge, this constitutes the first exhaustive, end-to-end application of LLM-based agents for structured oncology data extraction at scale
Traditional efforts toward automating the extraction of structured data from clinical text have relied heavily on rule-based systems or shallow machine learning models (e.g., Conditional Random Fields, Support Vector Machines), each requiring extensive domain-specific feature engineering [1][2][3][4]. In the last decade, however, transformerbased models [5], such as Bidirectional Encoder Representations from Transformers (BERT) [6] and its domain-specific variants like ClinicalBERT [7][8][9], have significantly improved upon classical approaches across a range of clinical natural language processing (NLP) tasks. More recently, the approach has shifted from fine-tuning domain specific models [10,11] to using the generalized abilities of frontier large language models (LLMs) like GPT-4 and GPT-5 [12,13] to extract key concepts from EHR records [14].
Nevertheless, prior works often assume relatively uniform data (such as a single cancer diagnosis or one type of note), single-document inputs, or a very limited set of concepts. Published studies have demonstrated entity extraction for discrete variables such as tumor stage from pathology reports [15,16], receptor or genomic biomarkers from pathology or genomic reports [17,18], and initial regimens or lines of therapy for select cancers [19,20]. However, these approaches extract variables independently without synthesizing findings across multiple documents to resolve contradictions or complete partial information. This study is distinct in that it targets patient-level synthesis across heterogeneous, longitudinal records, where oncology-specific concepts must be inferred by collating and contextualizing information scattered across multiple notes and data types, rather than relying on single-step extraction from a uniform document.
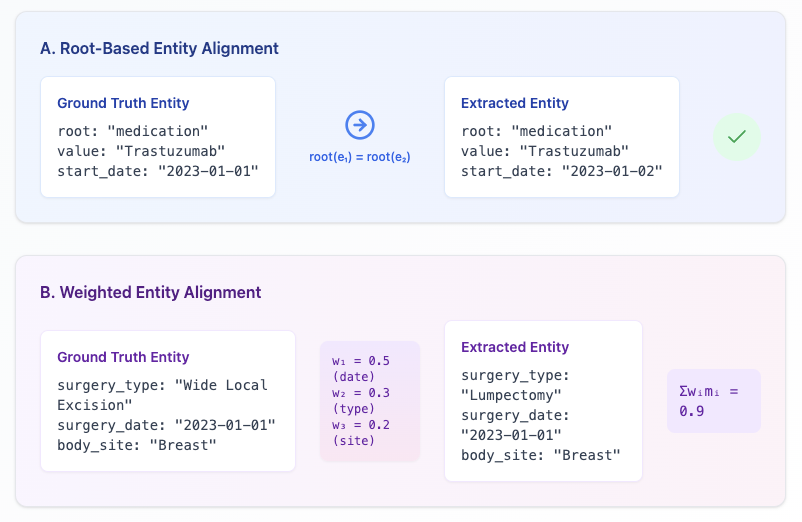
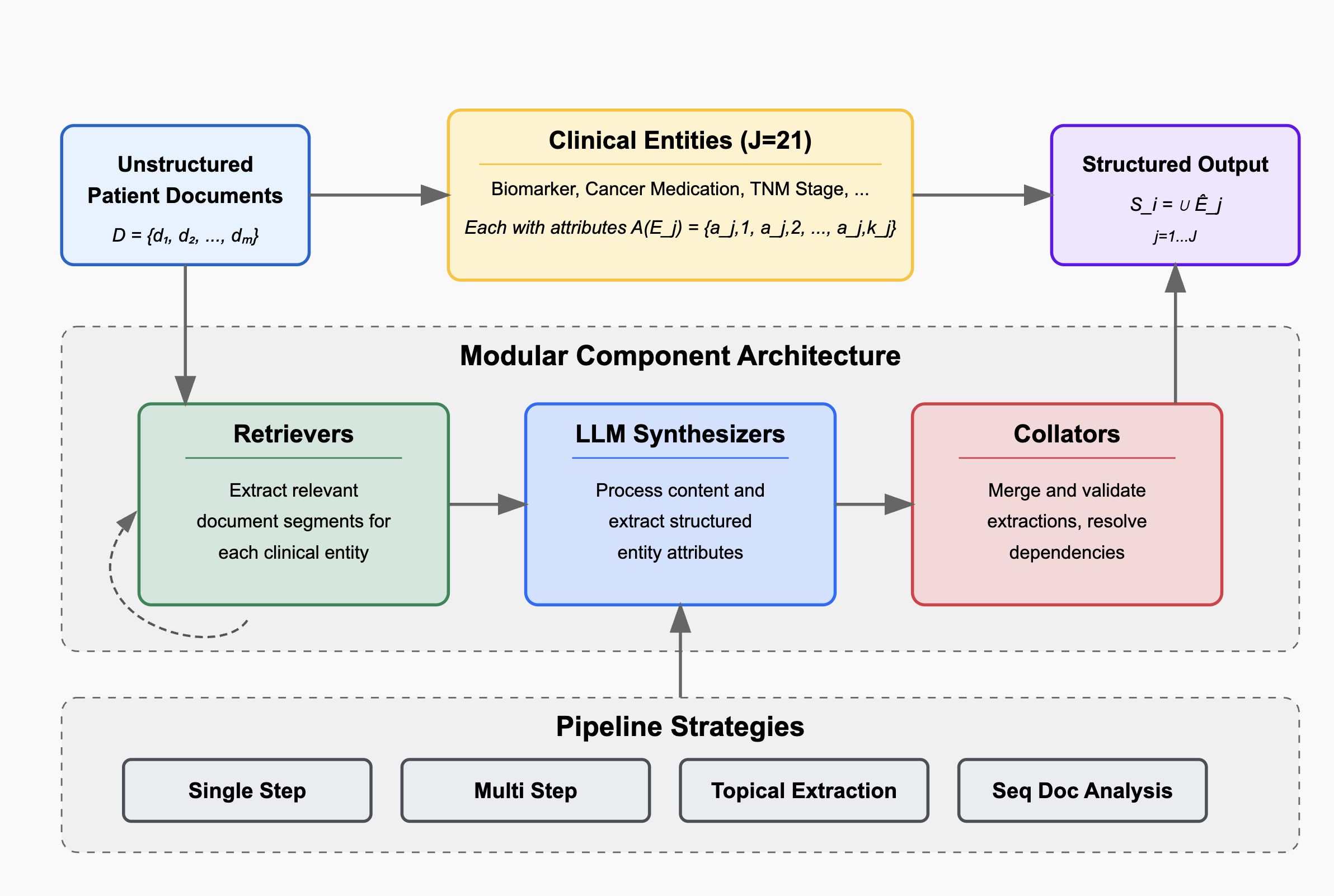
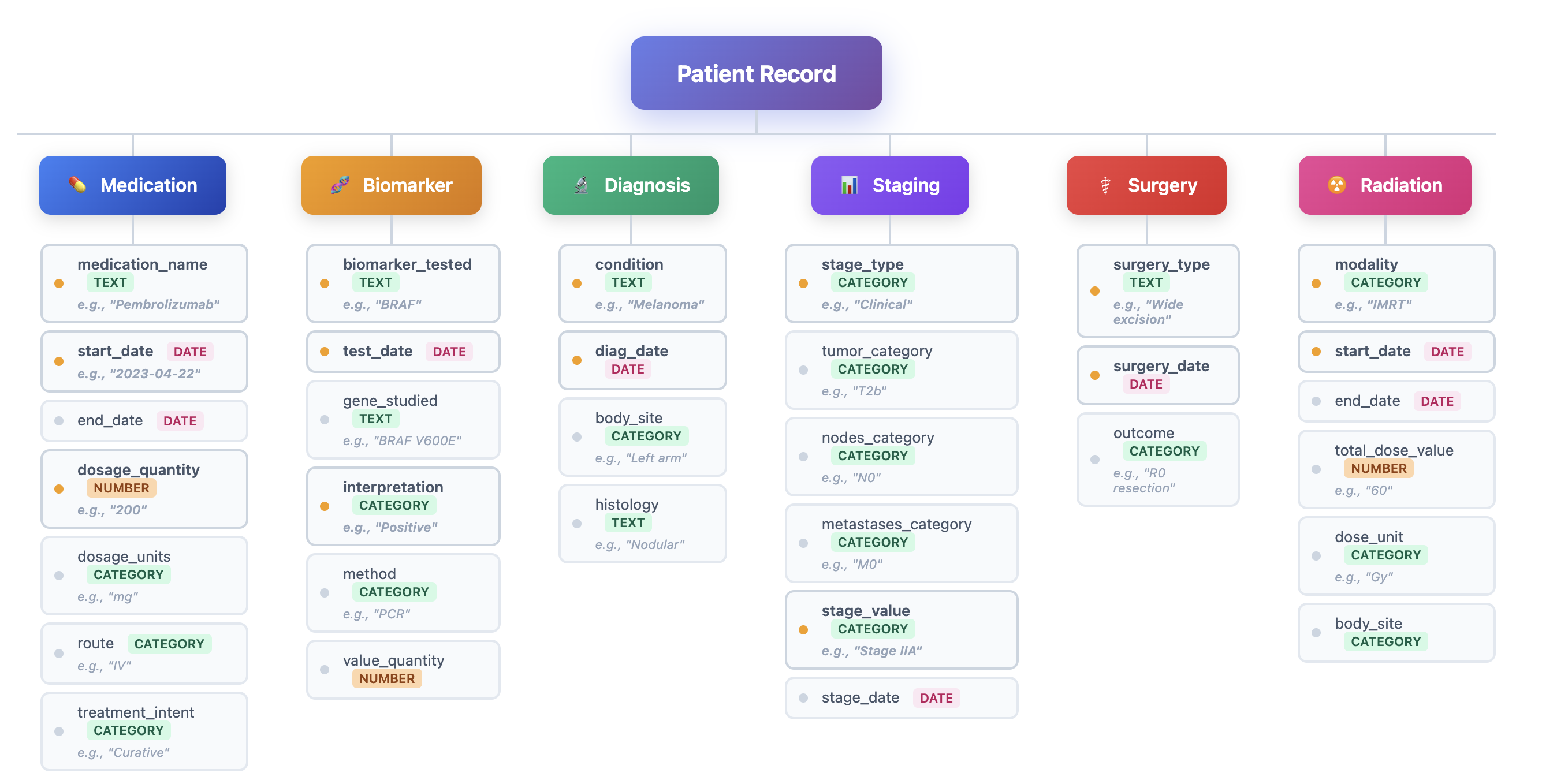
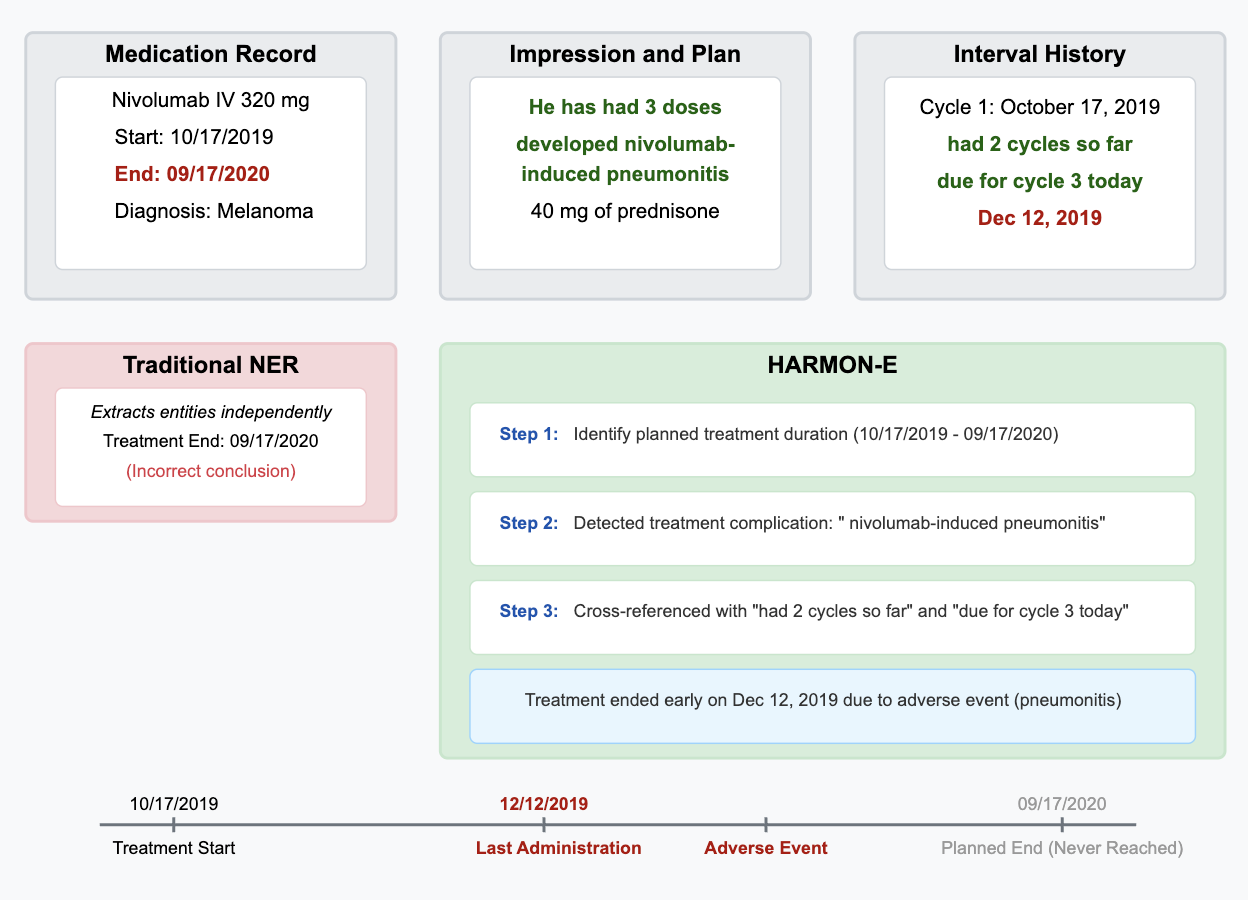
To address these challenges, we introduce HARMON-E : Hierarchical Agentic Reasoning for Multi-source Oncology Notes to Extract structured data Our framework systematically decomposes oncology data extraction into modular, iterative steps that leverage large language models (LLMs) as “reasoning agents” that interleave retrieval with multi-step inference to reconcile conflicting evidence, normalize temporal references, and derive implicit variables not explicitly stated in the text -such as inferring treatment discontinuation dates from adverse event narratives (Figure 1). Concretely, HARMON-E combines context-aware indexing, adaptive retrieval methods (including vector-based search and rule-based actions), and LLM-driven data synthesis into a unified workflow. By adopting a hierarchical, agentic approach, we mitigate common pitfalls in extracting complex, interdependent oncology data and achieve end-to-end alignment with standardized pre-defined set of clinical variables comprising 103 attributes across 16 entity types-including Biomarker, Medication, Diagnosis, Staging, Surgery and Radiation entities as outlined in Table 1 and Figure 3.
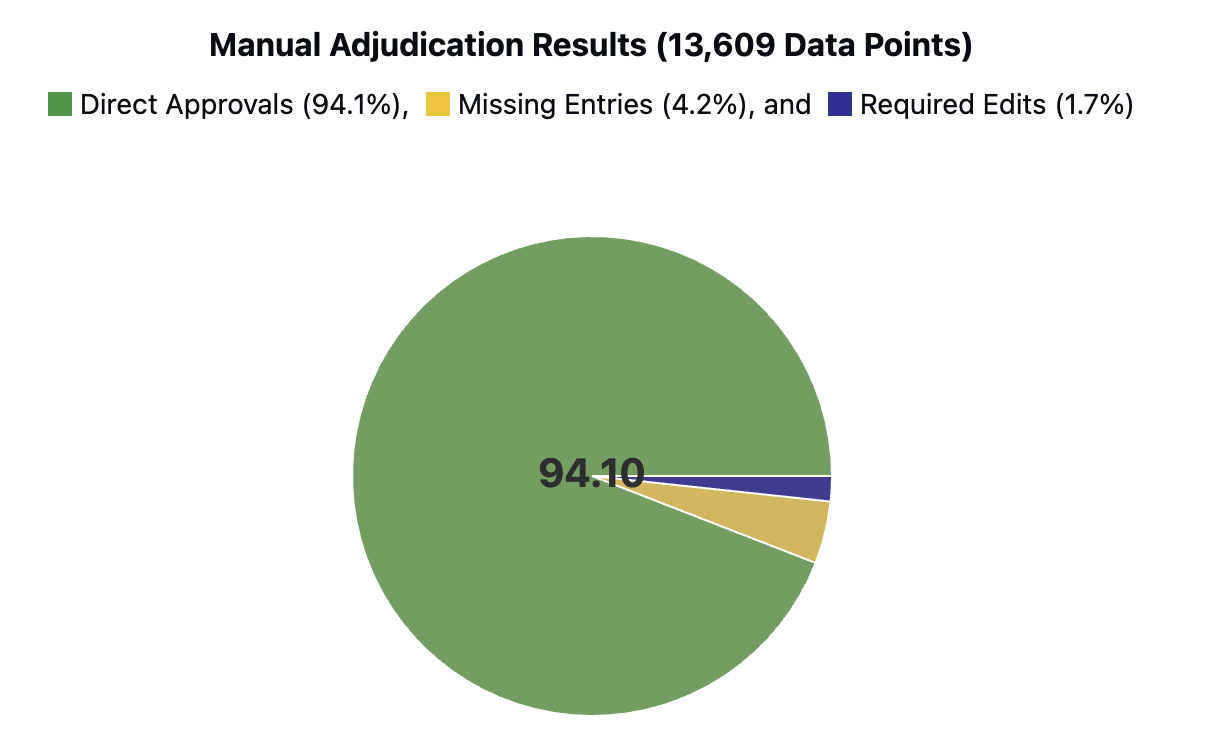
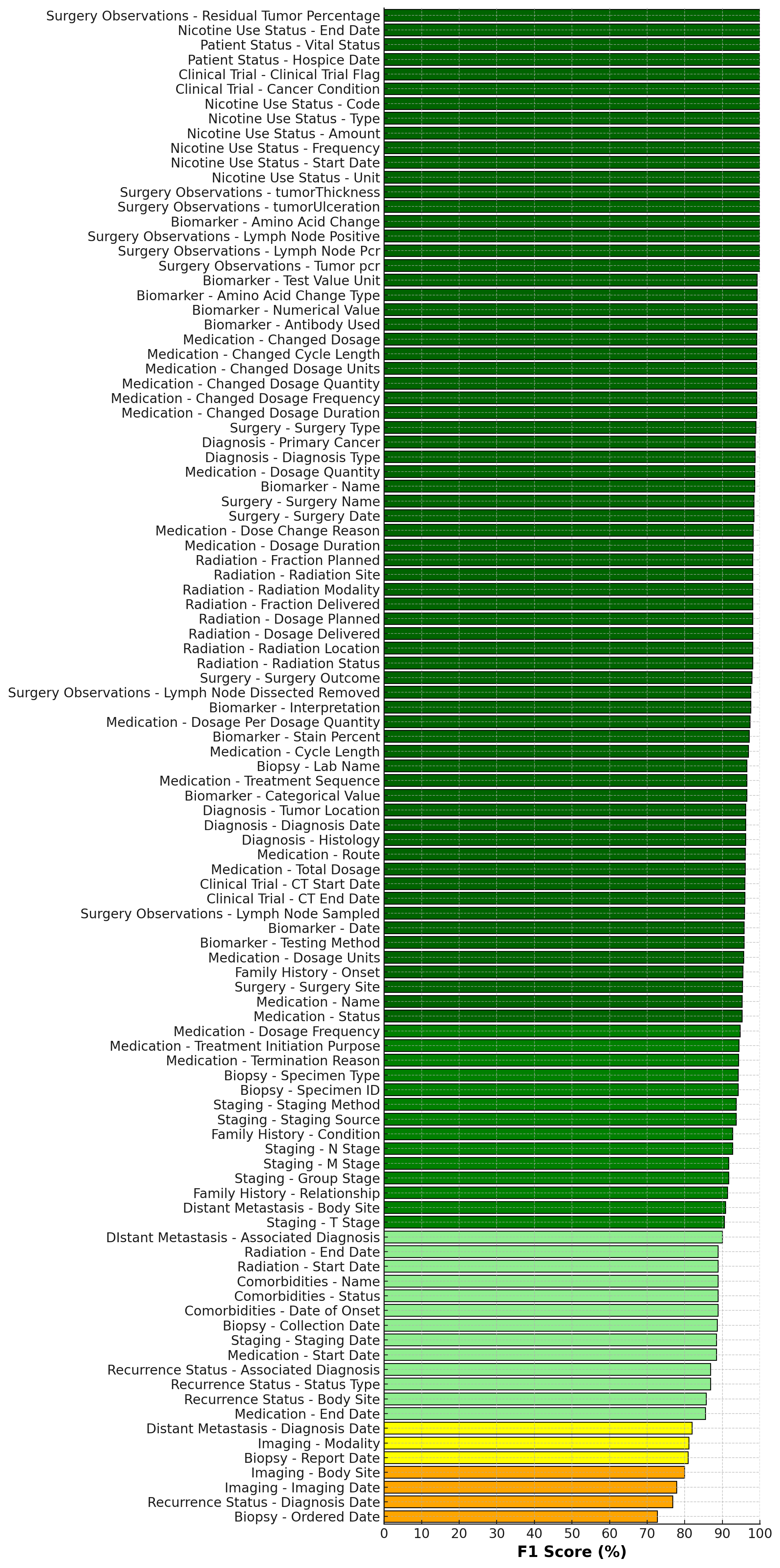
We validate HARMON-E on a large-scale real-world dataset of over 400,000 unstructured clinical notes and scanned PDFs belonging to 2,250 cancer patients. Our evaluation covers 103 oncology-specific variables-ranging from histopathological findings and biomarker statuses to treatment patterns and disease progression. HARMON-E consistently delivers high accuracy, with 100 out of 103 variables exceeding an F1score of 80%, and crucial fields such as biomarker assessments and medication data surpassing 95% accuracy. Beyond these metrics, when integrated into a data curation platform, our system demonstrates a direct manual approval rate of 94.1%, substantially reducing human annotation burden without sacrificing data quality.
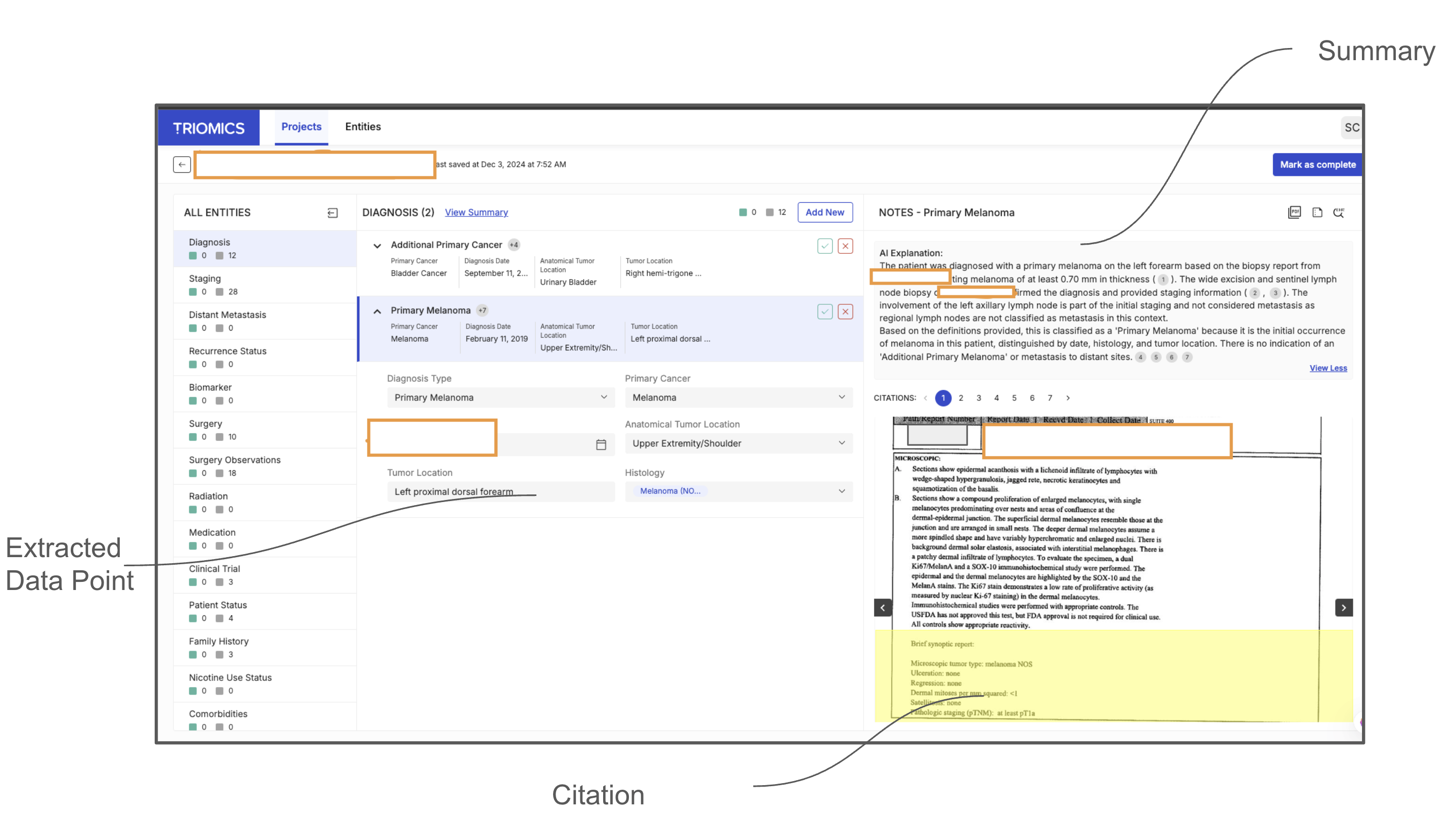
Our contributions can be summarized as follows -1. High-accuracy LLM-based agentic workflows. We provide the first evidence that large language models, organized into an agentic workflow, can exceed 95% F1-score on key oncology concepts at scale, validated against a corpus of 940,923 data points derived from 400,000 clinical notes covering 2,250 patients. 2. Real-world integration and user acceptance. We also demonstrate that when integrated into an interactive data curation platform, 94% of the extracted data points get directly accepted by professional oncology abstractors without modification, substantially reducing manual review time. 3. Novel evaluation framework. We propose an evaluation methodology which moves beyond traditional named-entity recognition metrics by aligning performance assessment with the real-world requirements of oncology data curation and quality monitoring.
Early Clinical NLP and Rule-Based Methods: Early clinical NLP relied primarily on rule-based systems, utilizing domain-specific lexicons and hand-crafted patterns [3,21,22]. These systems, though precise in certain scenarios, required substantial manual effort and were brittle when encountering variability [1,23]. Notable examples include MedXN for medication extraction [24], comprehensive clinical NL
This content is AI-processed based on open access ArXiv data.