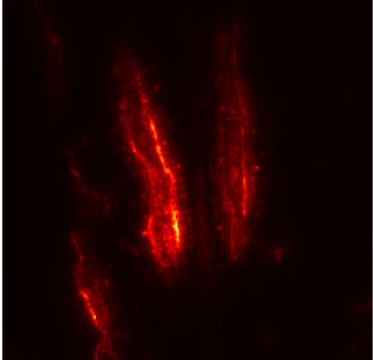
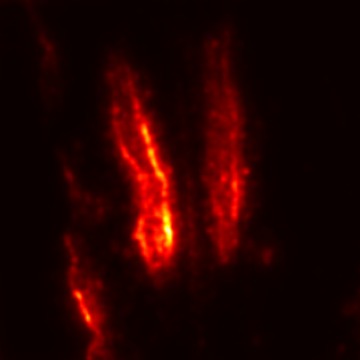
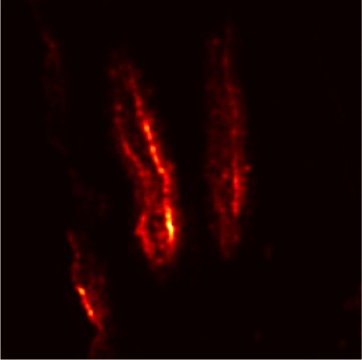
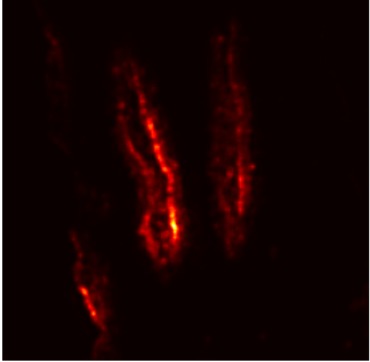
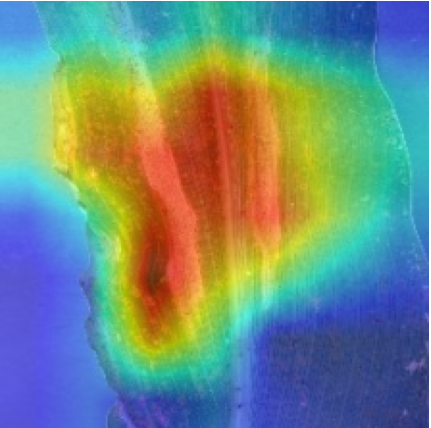
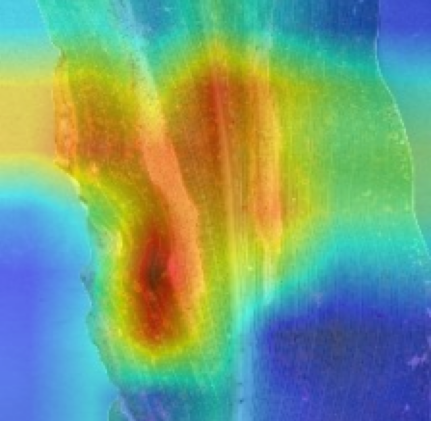
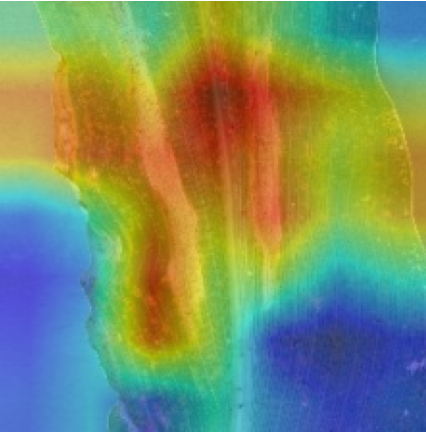
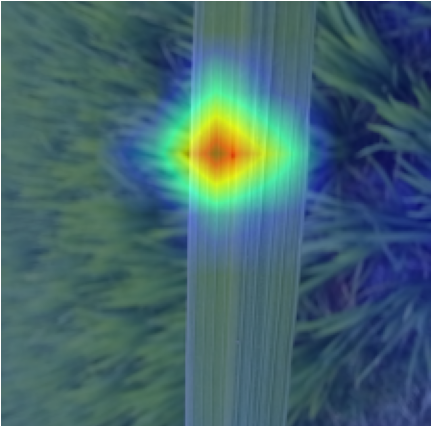
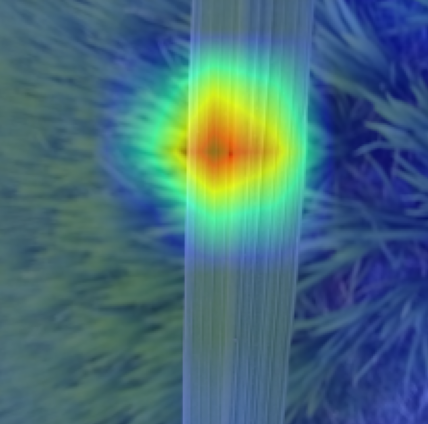
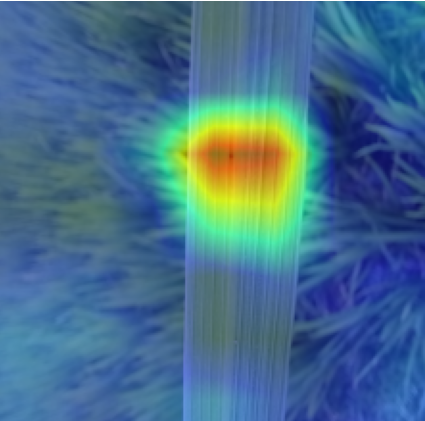
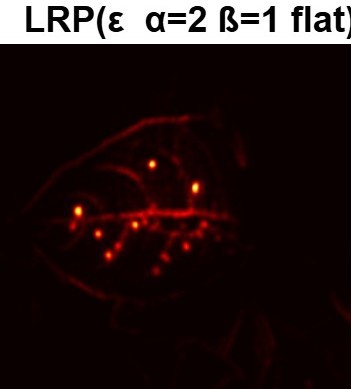
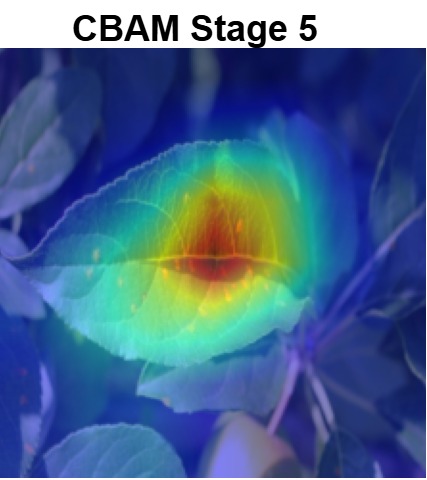
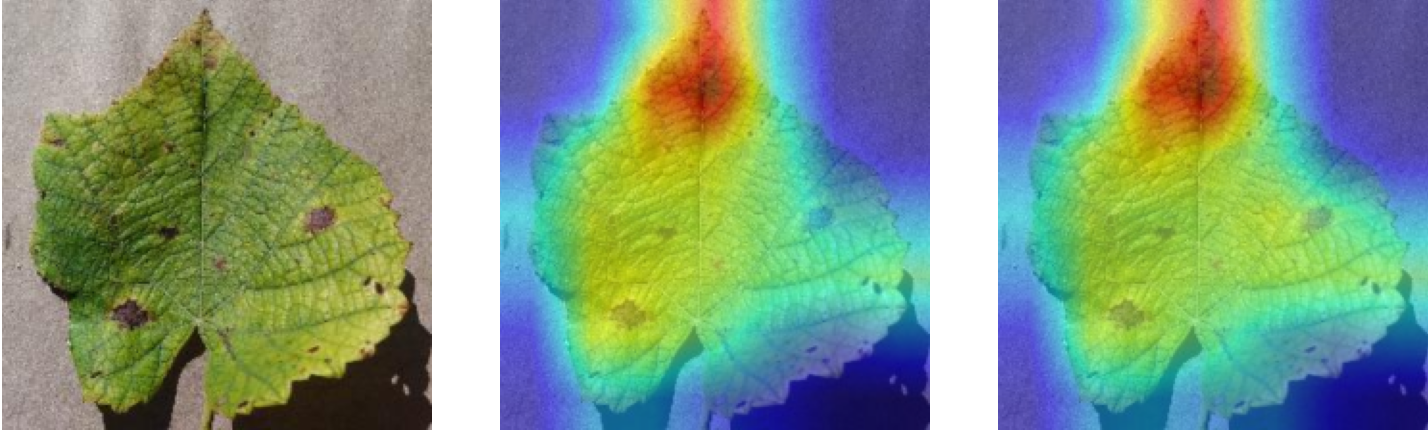
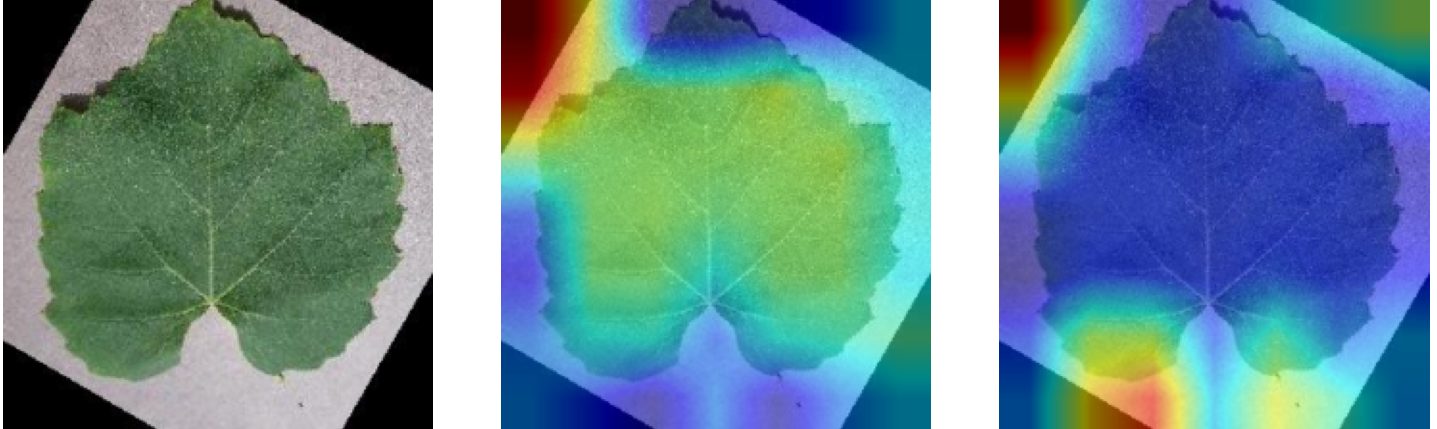
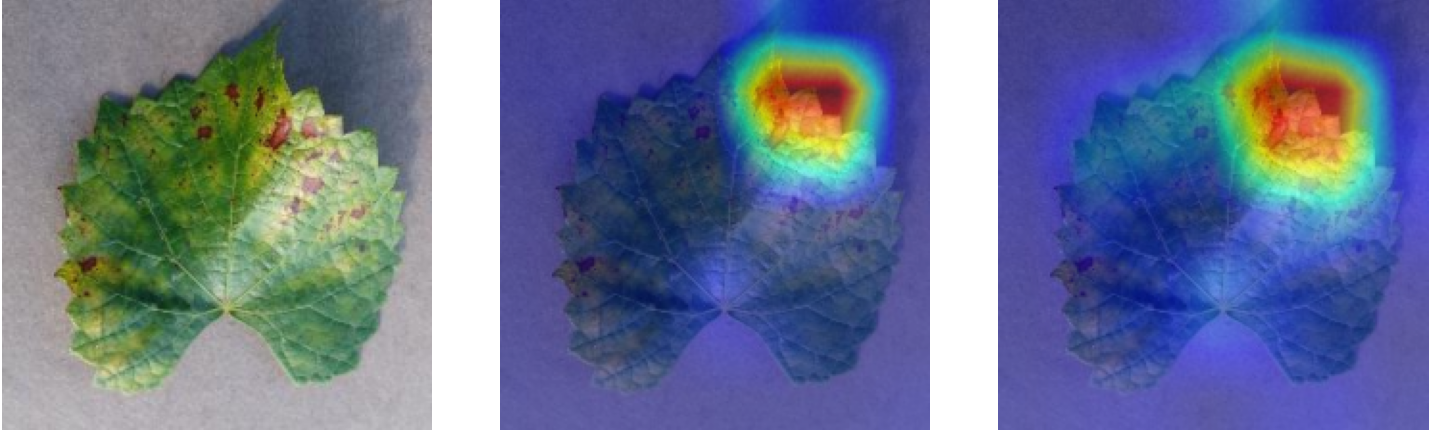
Plant diseases pose a significant threat to global food security, necessitating accurate and interpretable disease detection methods. This study introduces an interpretable attention-guided Convolutional Neural Network (CNN), CBAM-VGG16, for plant leaf disease detection. By integrating Convolution Block Attention Module (CBAM) at each convolutional stage, the model enhances feature extraction and disease localization. Trained on five diverse plant disease datasets, our approach outperforms recent techniques, achieving high accuracy (up to 98.87%) and demonstrating robust generalization. Here, we show the effectiveness of our method through comprehensive evaluation and interpretability analysis using CBAM attention maps, Grad-CAM, Grad-CAM++, and Layer-wise Relevance Propagation (LRP). This study advances the application of explainable AI in agricultural diagnostics, offering a transparent and reliable system for smart farming. The code of our proposed work is available at https://github.com/BS0111/PlantAttentionCBAM.
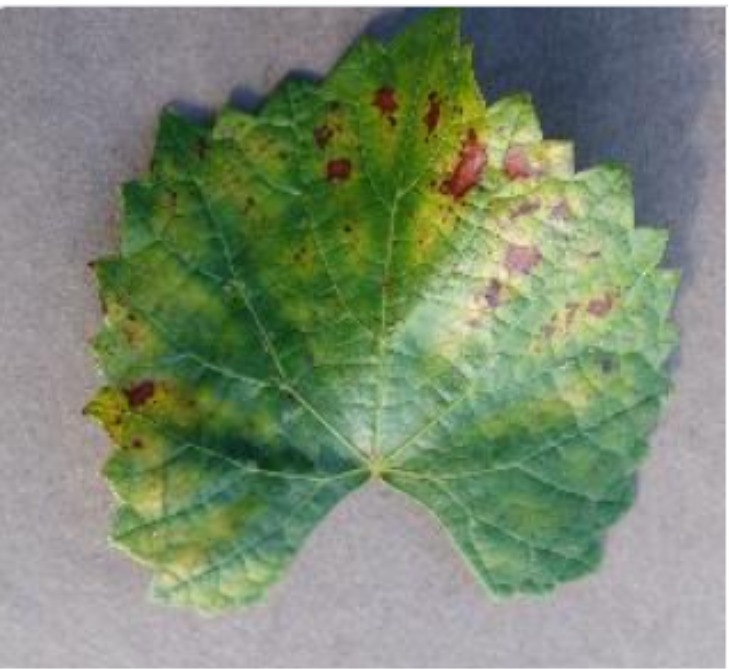
The agriculture industry is essential in maintaining food security worldwide, but different crops suffer from a variety of diseases due to varying weather conditions, posing a major threat to crop yield and quality. These diseases are often caused by factors such as extreme temperatures, microbial infections, and changes in humidity or soil conditions. Farmers have always relied on human examination to identify diseases, which is labour-intensive, prone to mistakes, and ineffective on large farms. As a result, among the most crucial areas of research in smart agriculture is the automation of plant disease identification and classification utilizing various Artificial Intelligence (AI) approaches [1,2]. In recent years, Convolutional Neural Networks (CNNs) based models in agriculture have performed well on challenges such as classifying plant diseases. However, despite their effectiveness, CNNs often operate as black boxes, offering limited transparency into how predictions are made, hampering trust and broader adoption.
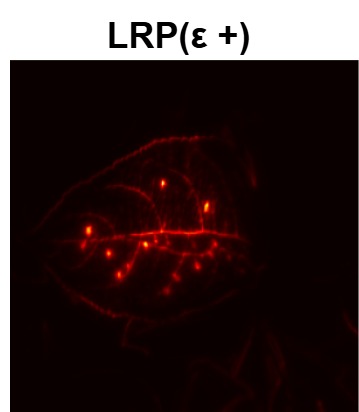
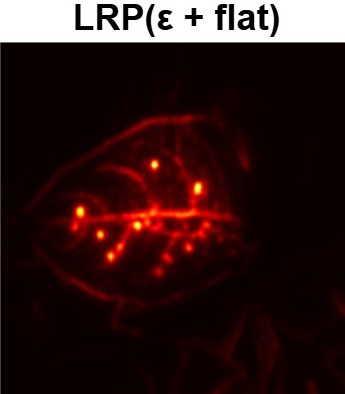
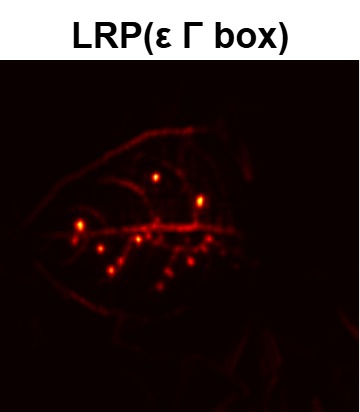
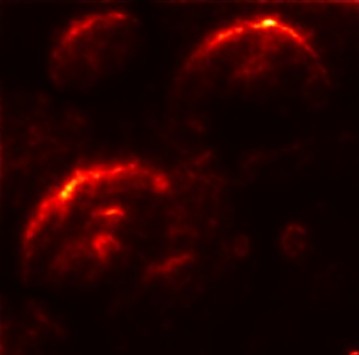
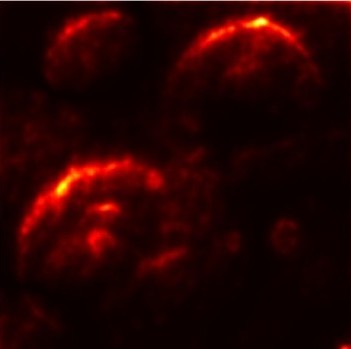
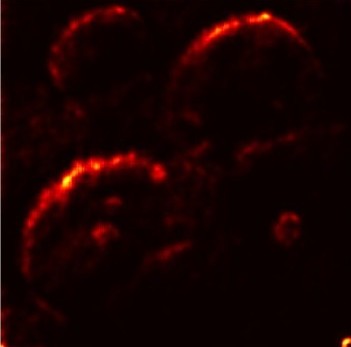
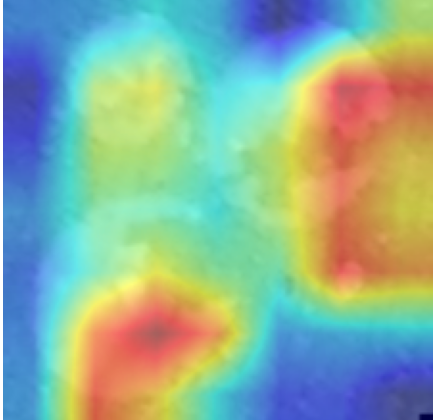
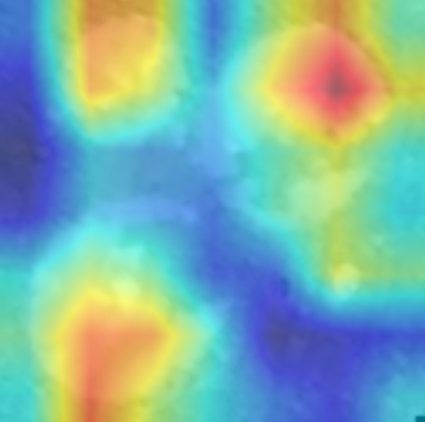
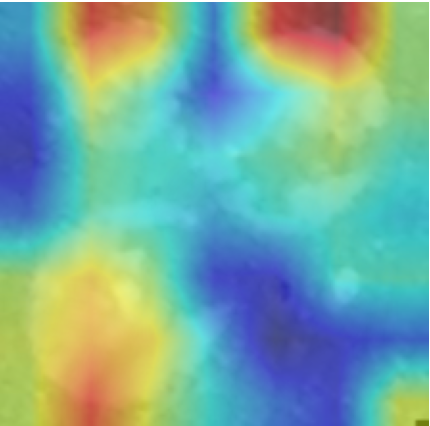
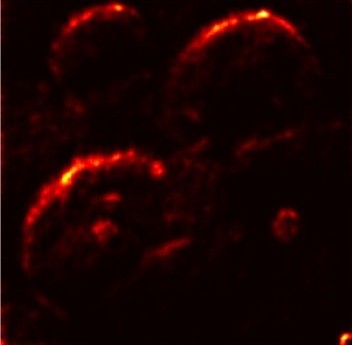
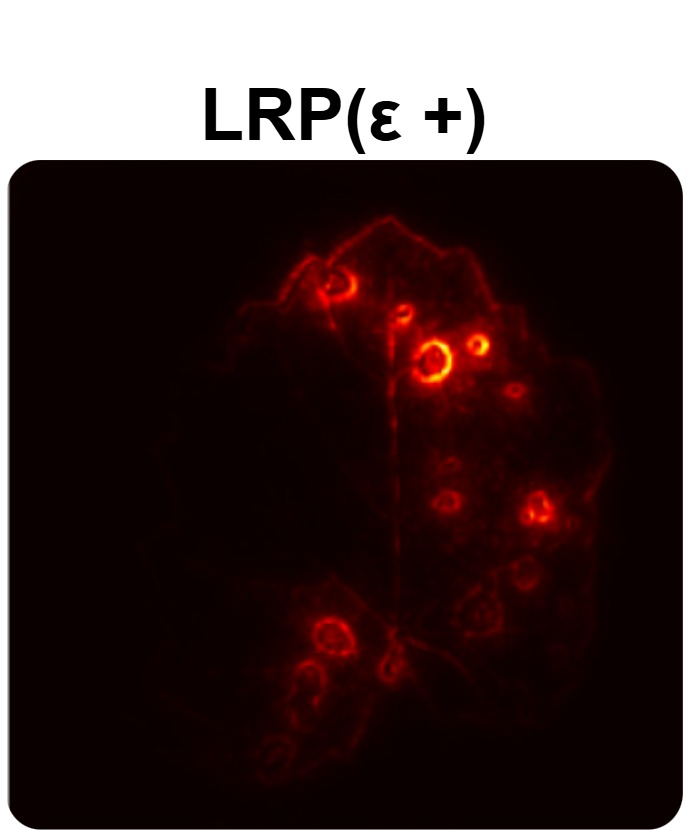
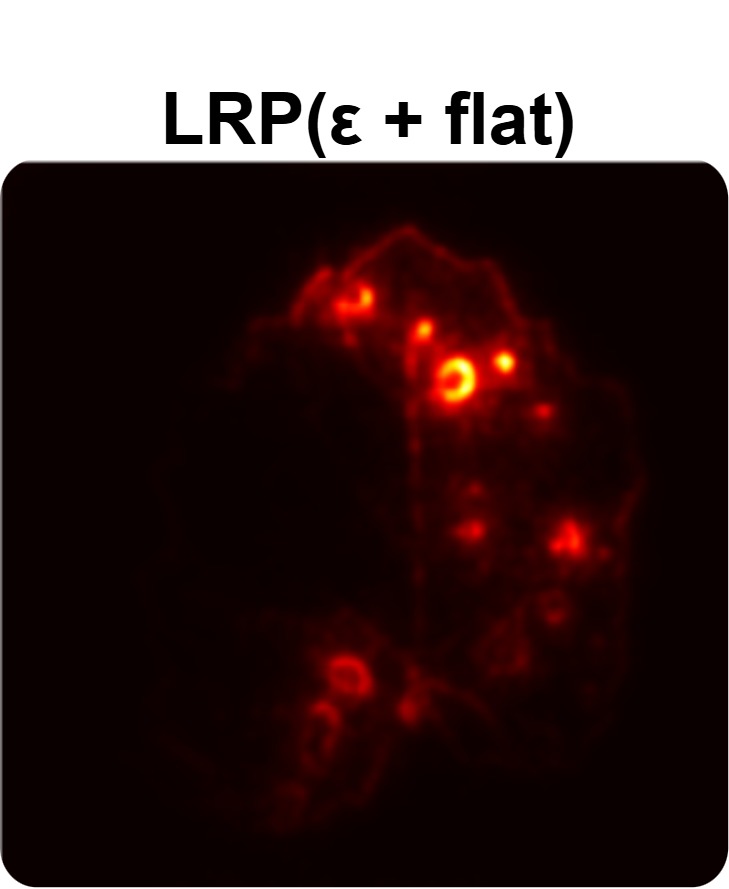
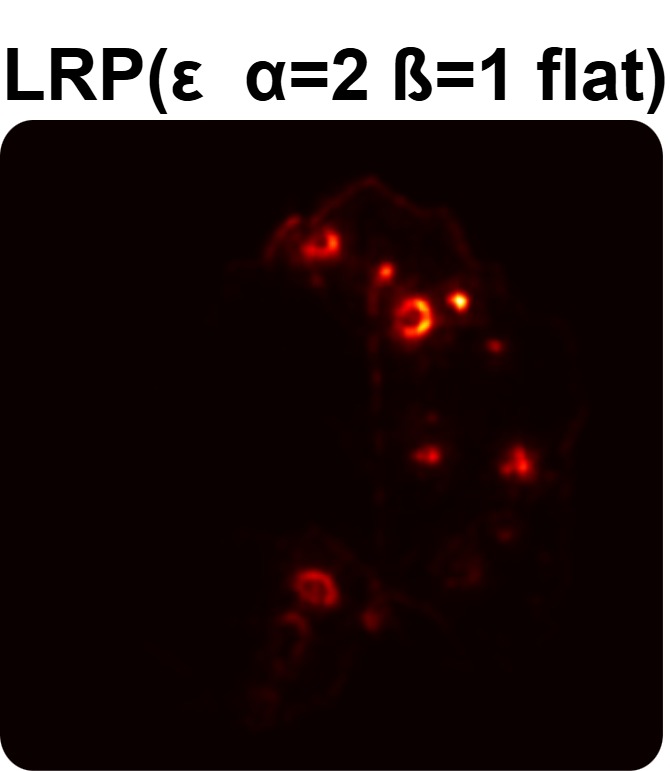
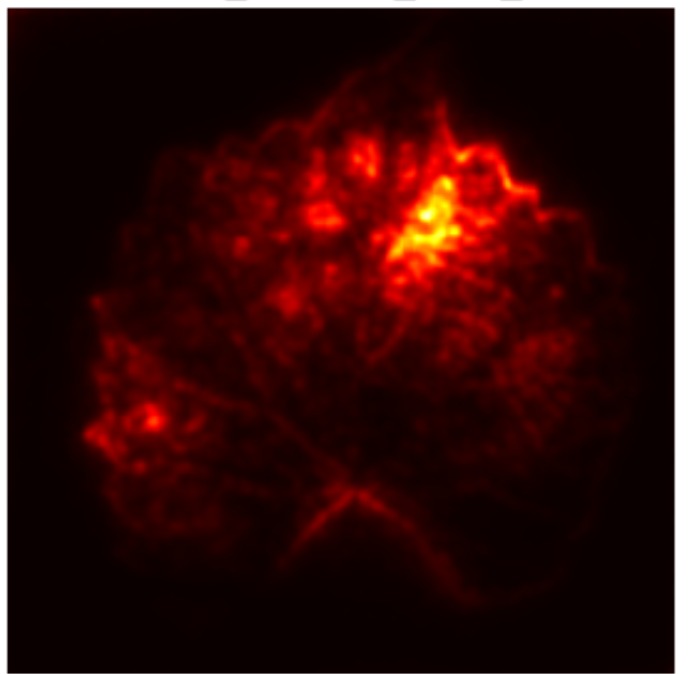
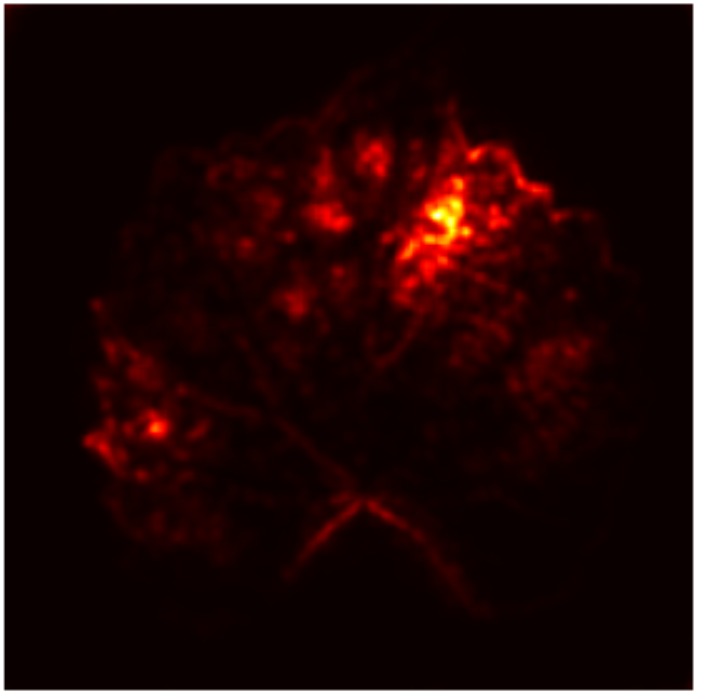
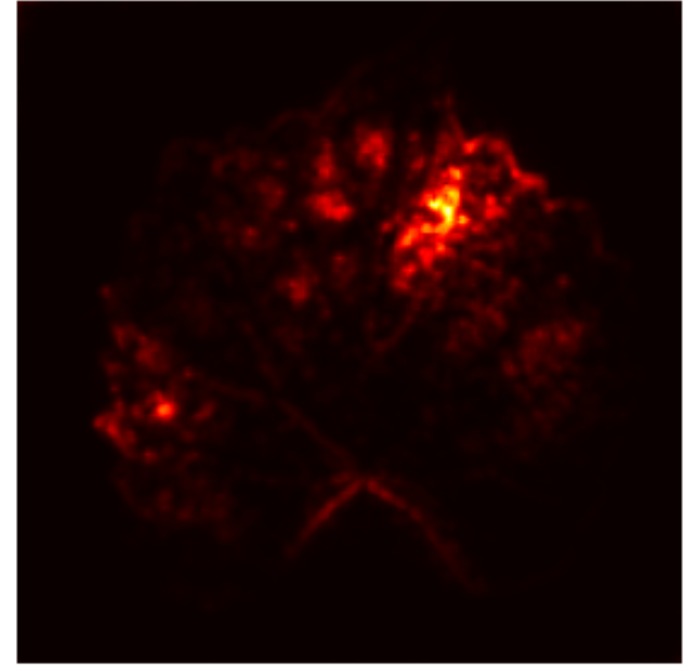
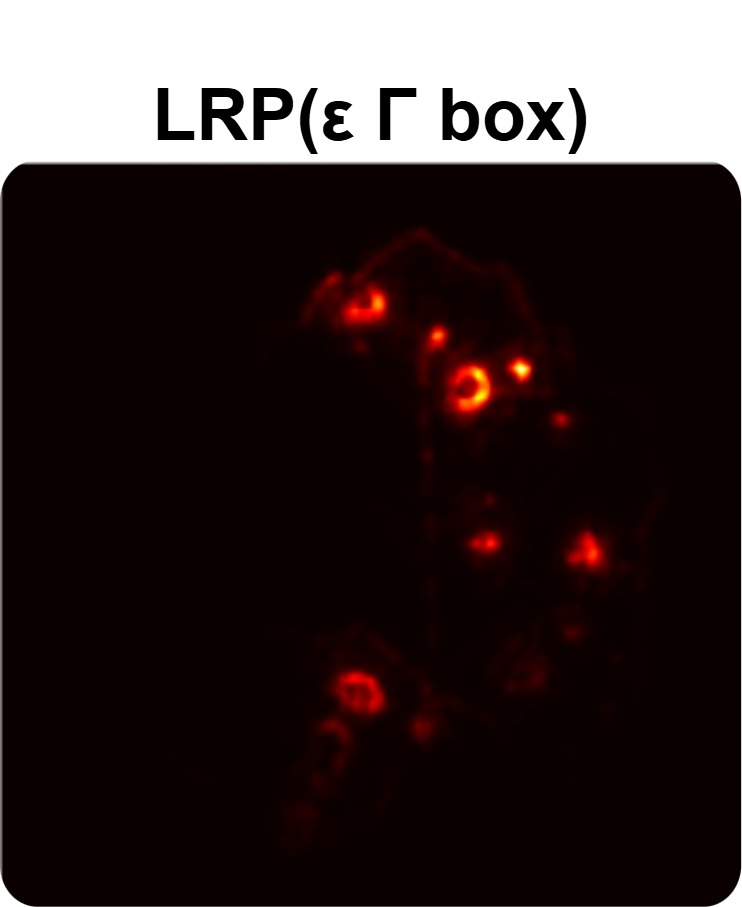
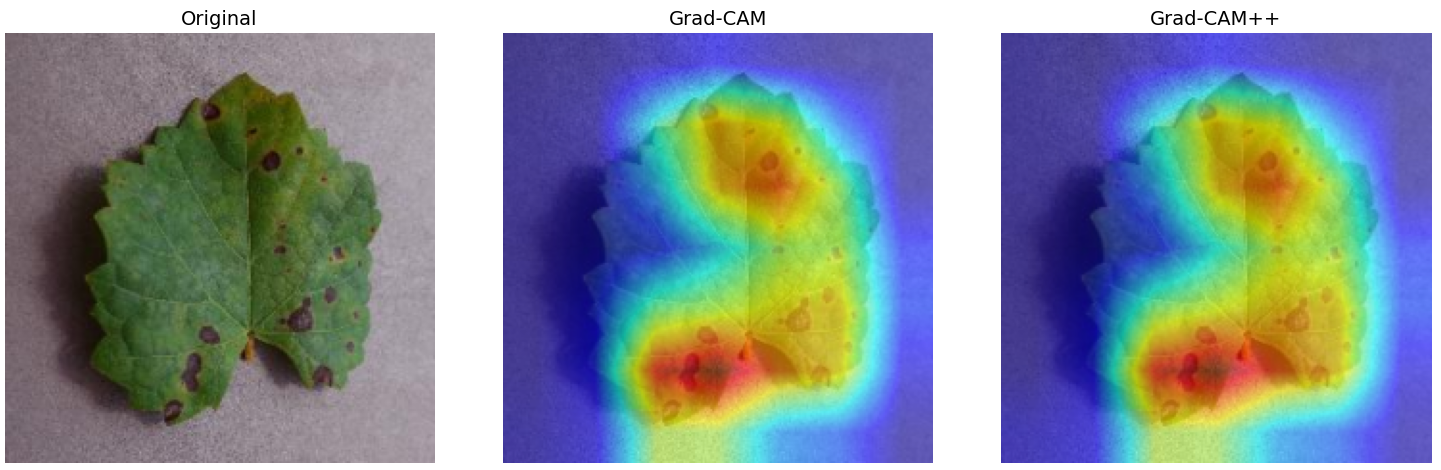
In order to tackle this, Explainable AI (XAI) methods have been developed enabling visual interpretation of model decisions through attention maps, GradCAM [3], and LRP [4]. While GradCAM and GradCAM++ [5] generate classdiscriminative localization maps using gradients from the final convolutional layers, they may lack resolution and be susceptible to noisy activations. LRP, in contrast, provides pixel-level attributions by backpropagating the model output through a set of layer-specific relevance propagation rules. This offers a more granular explanation of predictions, which is crucial in medical and agricultural diagnostics.
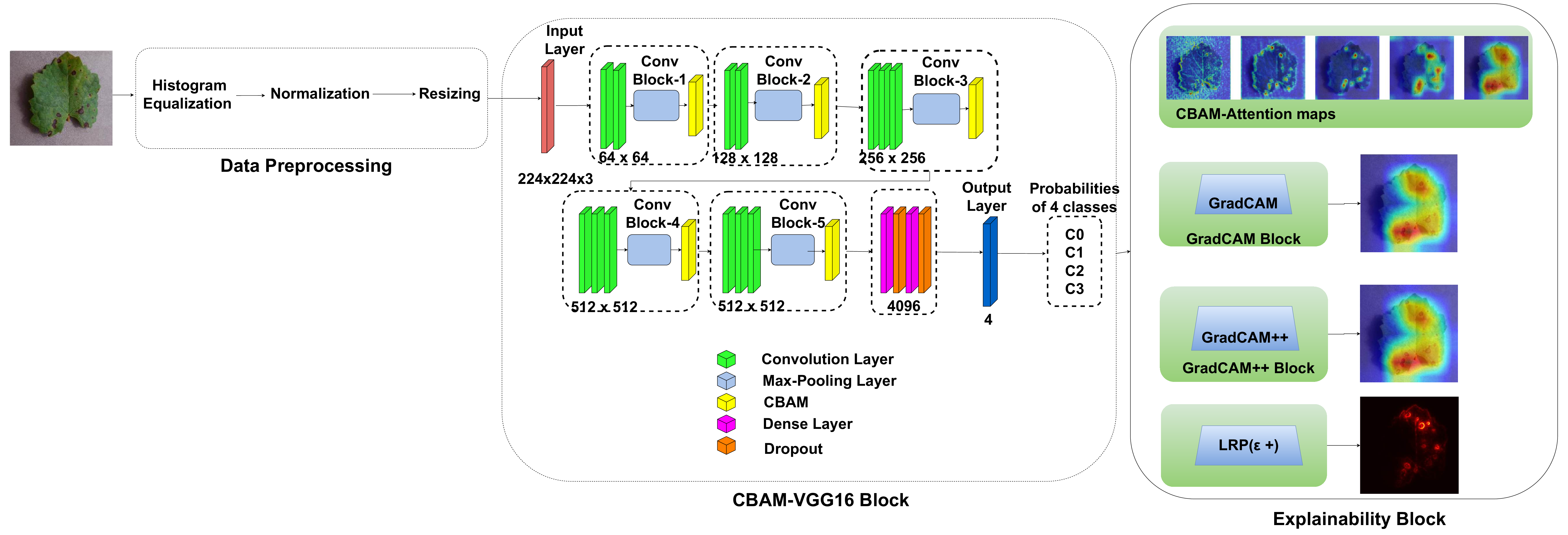
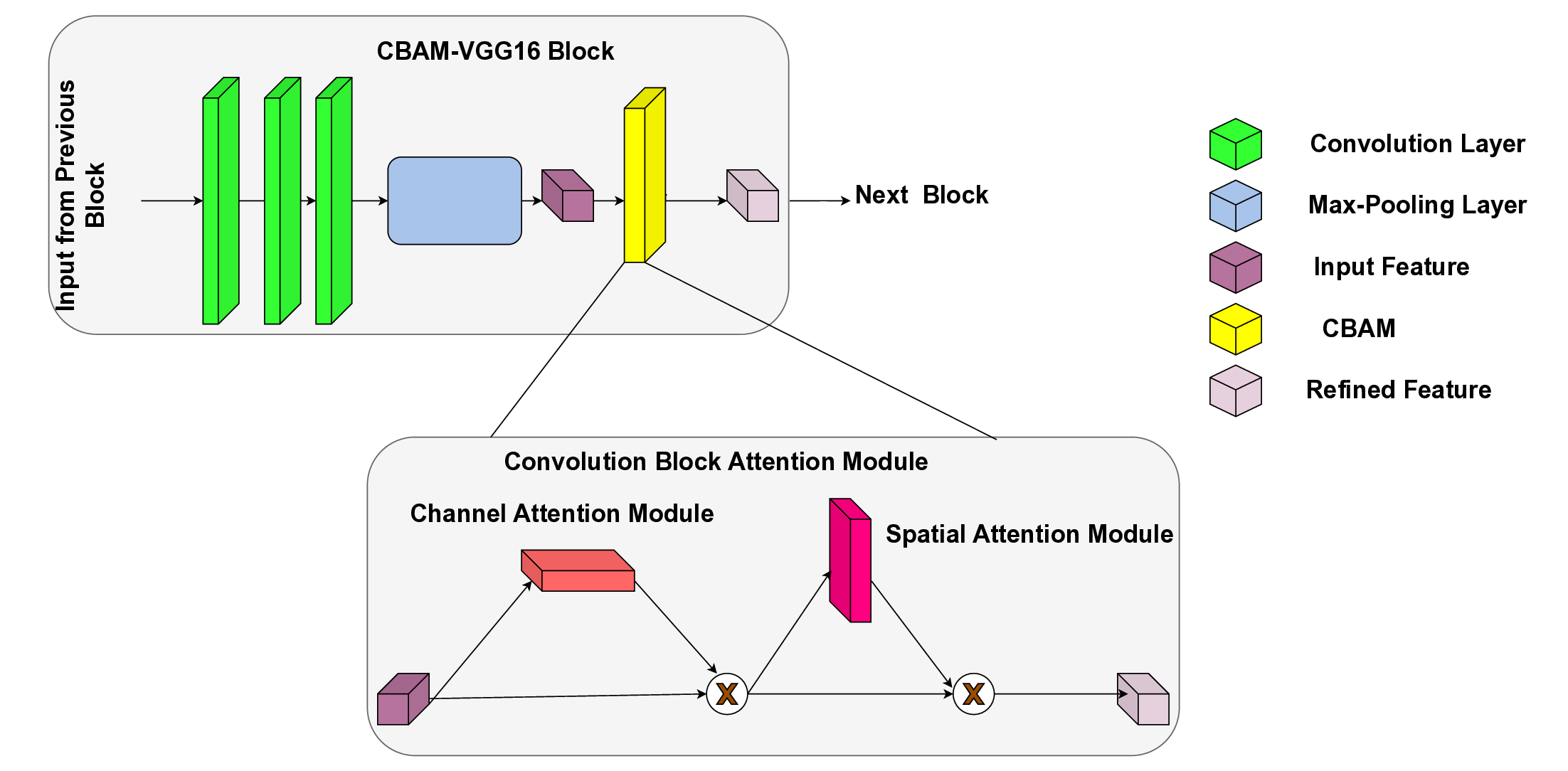
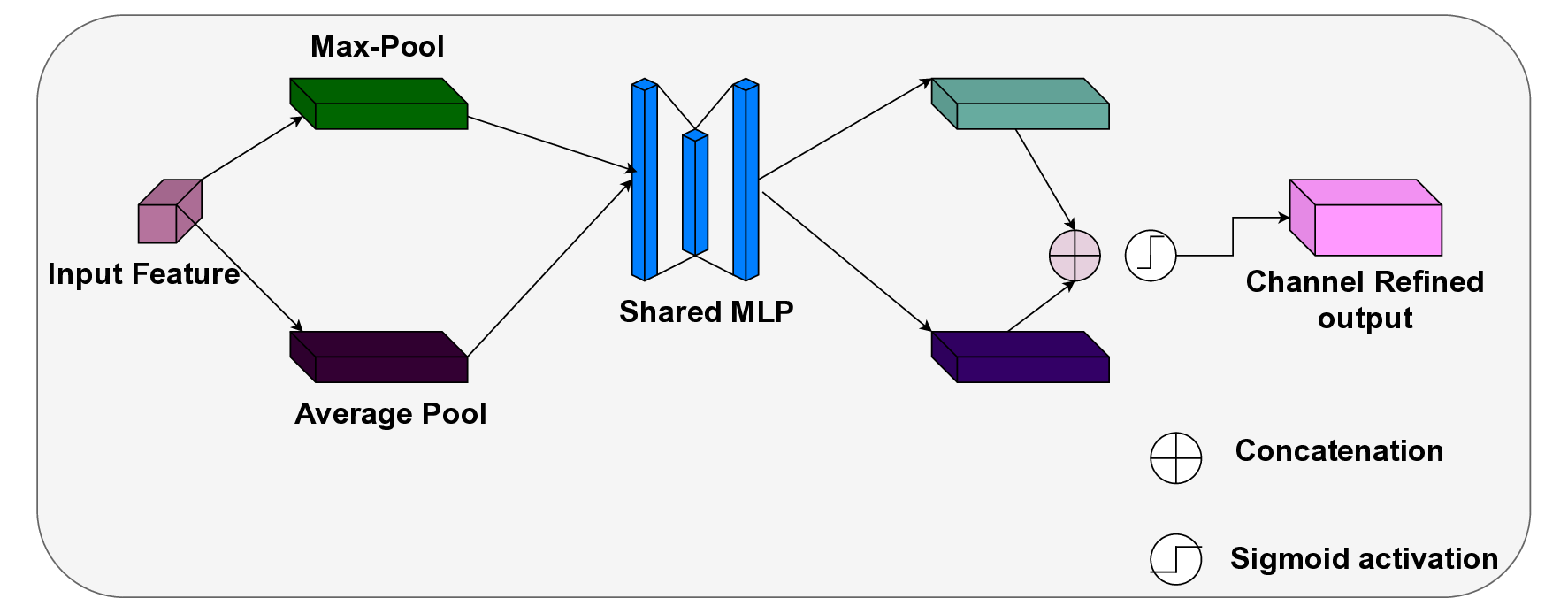
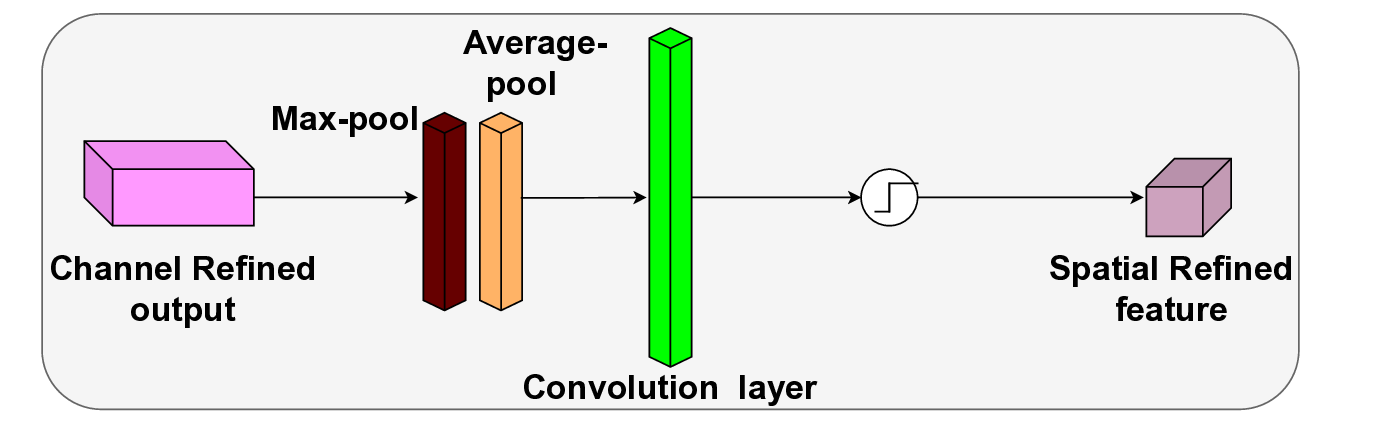
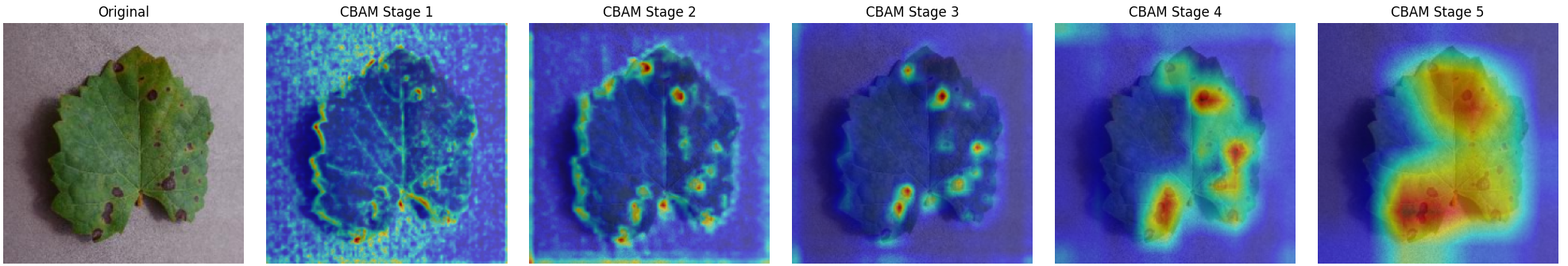
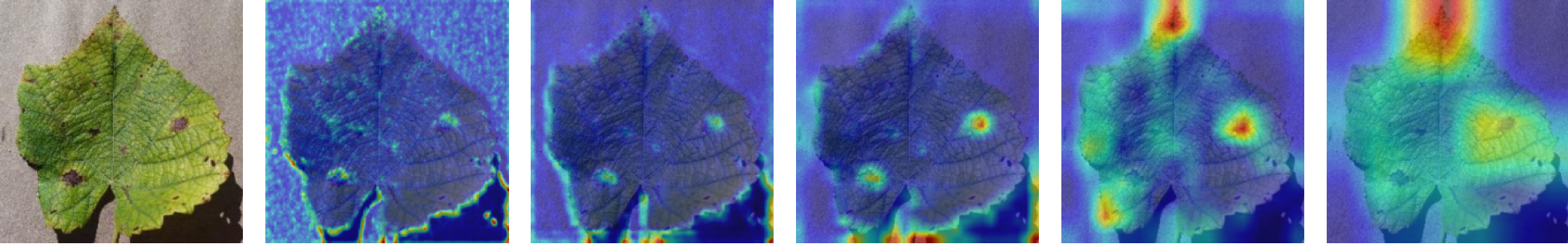
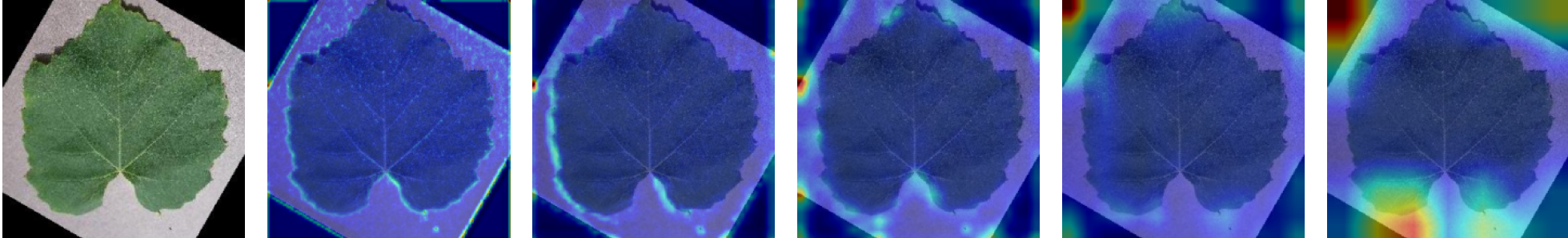
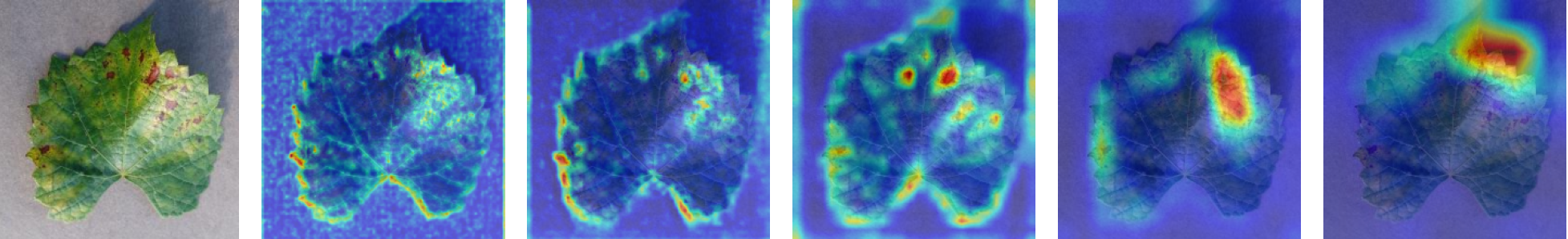
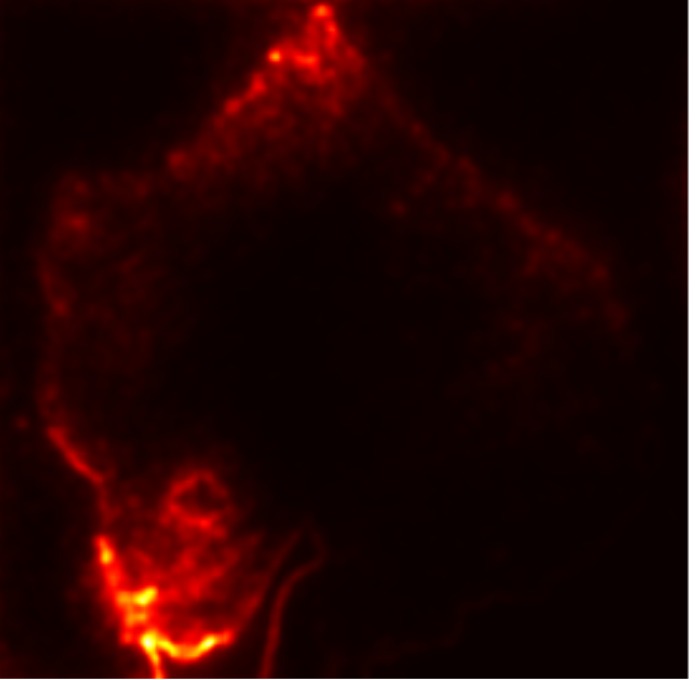
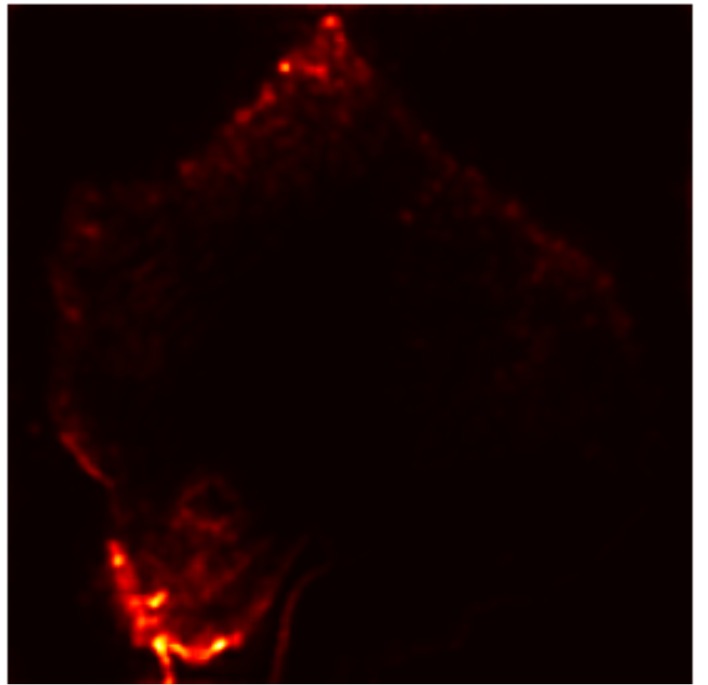
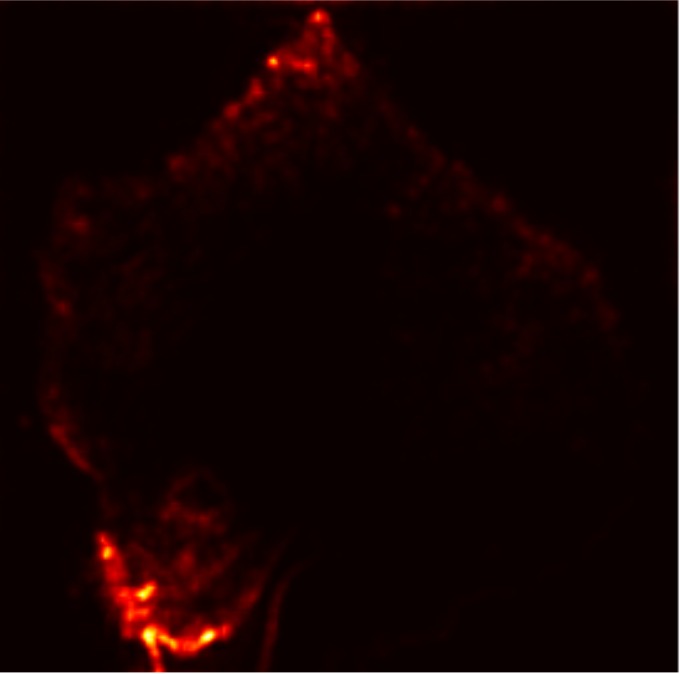
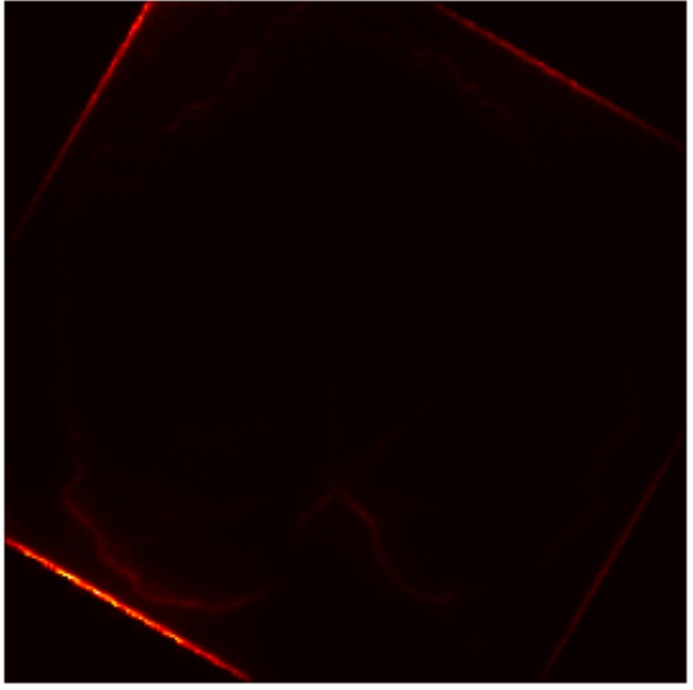
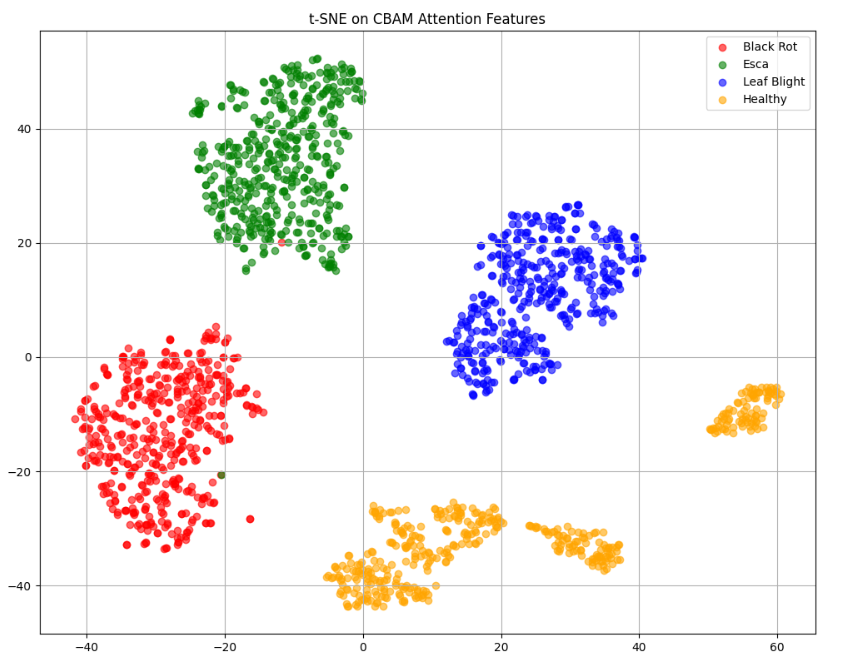
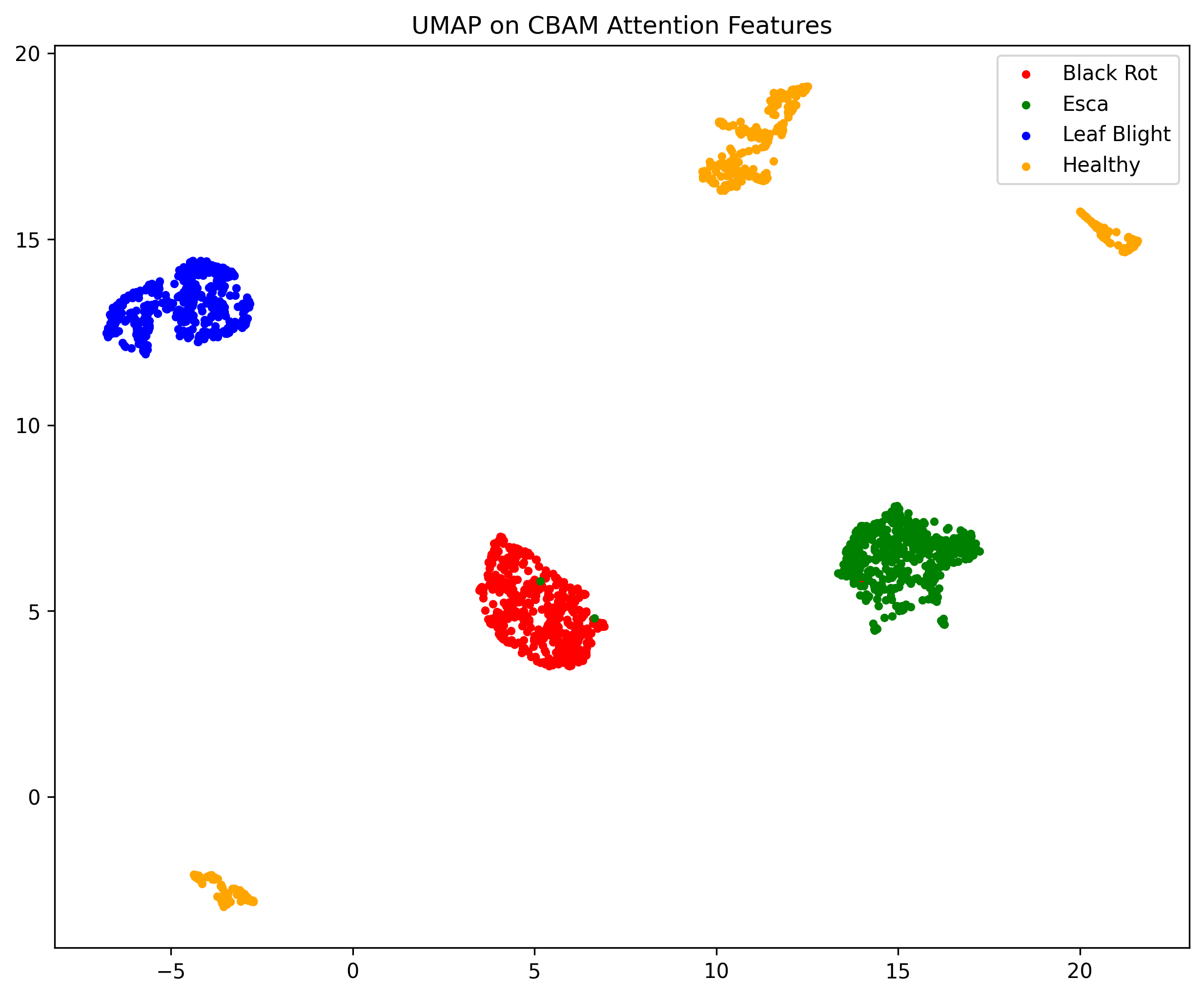
In this work, an explainable deep learning approach is proposed for the detection of plant leaf disease. Our architecture is based on the VGG16 [6] backbone enhanced with the CBAM [7] which introduces attention layers for inherent interpretability of the model with an emphasis on the most relevant features at both the channel and spatial levels. Following each of the five convolutional layers, CBAM modules are added to improve classification accuracy and localization of relevant features by capturing both spatial and channel-wise attention. Five distinct datasets are used to train the model, namely Apple, Plant Village, Embrapa, Maize and Rice to ensure the generalizability and applicability of our proposed method across diverse set of crops. Apart from the inherent interpretability of the proposed method’s decision-making process through CBAM layers, we also demonstrate interpretability using advanced explainability methods like LRP, Grad-CAM, and Grad-CAM++. We have also employed high-dimensional feature visualization techniques such as t-distributed Stochastic Neighbor Embedding (t-SNE) and Uniform Manifold Approximation and Projection (UMAP) to visualize feature in lower dimension for visualization of extracted features. Overall, this study advances the application of XAI for agricultural use by presenting an interpretable and performance-robust framework for plant disease classification. The following are thes main contributions of the proposed work. [11] has proposed LeafGAN architecture for the augmentation of diseased leaf images via transformation for improving the plant disease diagnosis system using large scale dataset. To solve the data imbalance problem of health vs unhealthy images Zhao et al. [12] have used a DoubleGAN architecture. Their architecture uses Wasserstein Generative Adversarial Networks (WGAN) and Superresolution Generative Adversarial Network (SRGAN) to balance the dataset. Some of the recent methods also used the fuzzy rank-based ensemble along with the pretrained CNNs [13] and fuzzy feature extraction [14] for the plant leaf detection.
Although methods like GradCAM [15] and LRP are commonly utilised to interpret CNN outputs, their application in detecting plant leaf diseases is still relatively new. In the context of plant disease identification, LRP has been utilised to highlight significant regions of a leaf, aiding in the detection of disease symptoms and affected zones [16]. To improve interpretability, a number of studies have looked into combining Grad-CAM using additional methods of explanation with the goal of addressing GradCAM’s shortcomings, including its tendency to produce noisy or vague visual outputs. Despite progress, issues like unclear GradCAM explanations and the complexity of LRP visualisations remain unresolved. A detailed survey of recent advances in plant disease detection is provided by Qadri et al. [17]. They have highlighted the issue and challenges in plant leaf disease detection using machine learning and deep learning based solutions.
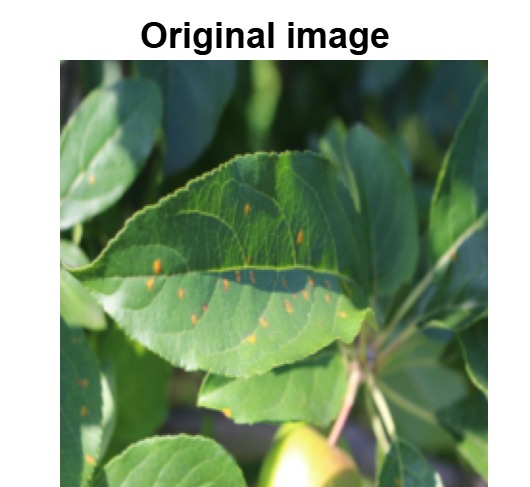
The overview of interpretable architecture used for identifying plant leaf disease is provided in Fig. 1. The input plant images undergo through data preprocessing to improve quality and guarantee alignment with the input structure of the proposed CBAM-VGG16 model. This invo
This content is AI-processed based on open access ArXiv data.