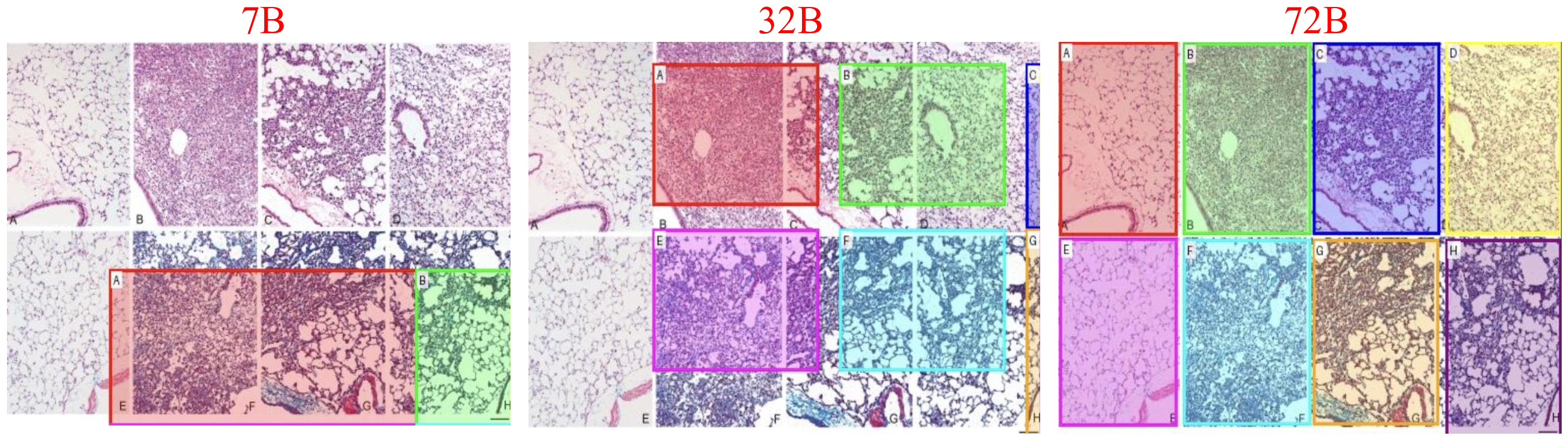
There is a growing interest in developing strong biomedical vision-language models. A popular approach to achieve robust representations is to use web-scale scientific data. However, current biomedical vision-language pretraining typically compresses rich scientific figures and text into coarse figure-level pairs, discarding the fine-grained correspondences that clinicians actually rely on when zooming into local structures. To tackle this issue, we introduce Panel2Patch, a novel data pipeline that mines hierarchical structure from existing biomedical scientific literature, i.e., multi-panel, marker-heavy figures and their surrounding text, and converts them into multi-granular supervision. Given scientific figures and captions, Panel2Patch parses layouts, panels, and visual markers, then constructs hierarchical aligned vision-language pairs at the figure, panel, and patch levels, preserving local semantics instead of treating each figure as a single data sample. Built on this hierarchical corpus, we develop a granularity-aware pretraining strategy that unifies heterogeneous objectives from coarse didactic descriptions to fine region-focused phrases. By applying Panel2Patch to only a small set of the literature figures, we extract far more effective supervision than prior pipelines, enabling substantially better performance with less pretraining data.
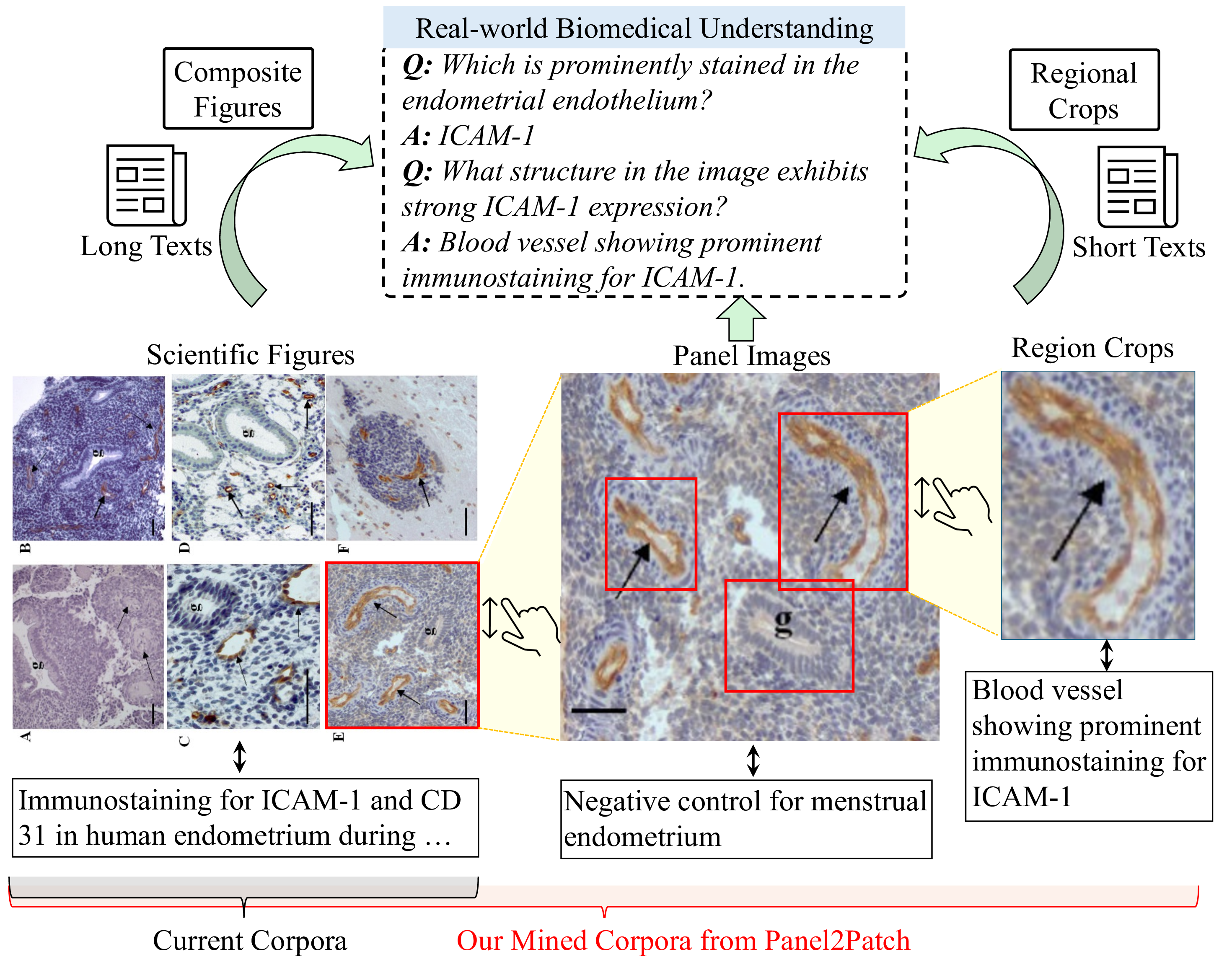
Generalist vision-language foundation models are rapidly becoming central to biomedical research by learning unified representations across radiology [4], cellular microscopy [44], and surgical videos [41,42] from largescale image-text pairs. A widely adopted strategy for constructing supervision is inspired by how human experts are trained: exposure to textbooks and educational articles that provide rich multimodal content capturing expert knowl-Figures Long Texts edge, visual patterns, and their semantic associations, enabling strong generalization to tasks such as visual question answering [21] and image captioning [9]. However, these models often struggle when tasks require fine-grained discrimination between similar attributes, spatial configurations, or subtle semantic distinctions. This gap stems from the nature of scientific corpora, where figures are mostly multi-panel and captions provide high-level summaries, leading to coarse figure-level pairs for pretraining [16,18,26] that favor thematic alignment but lack the localized grounding clinicians rely on when zooming into specific structures, as illustrated in Fig. 1.
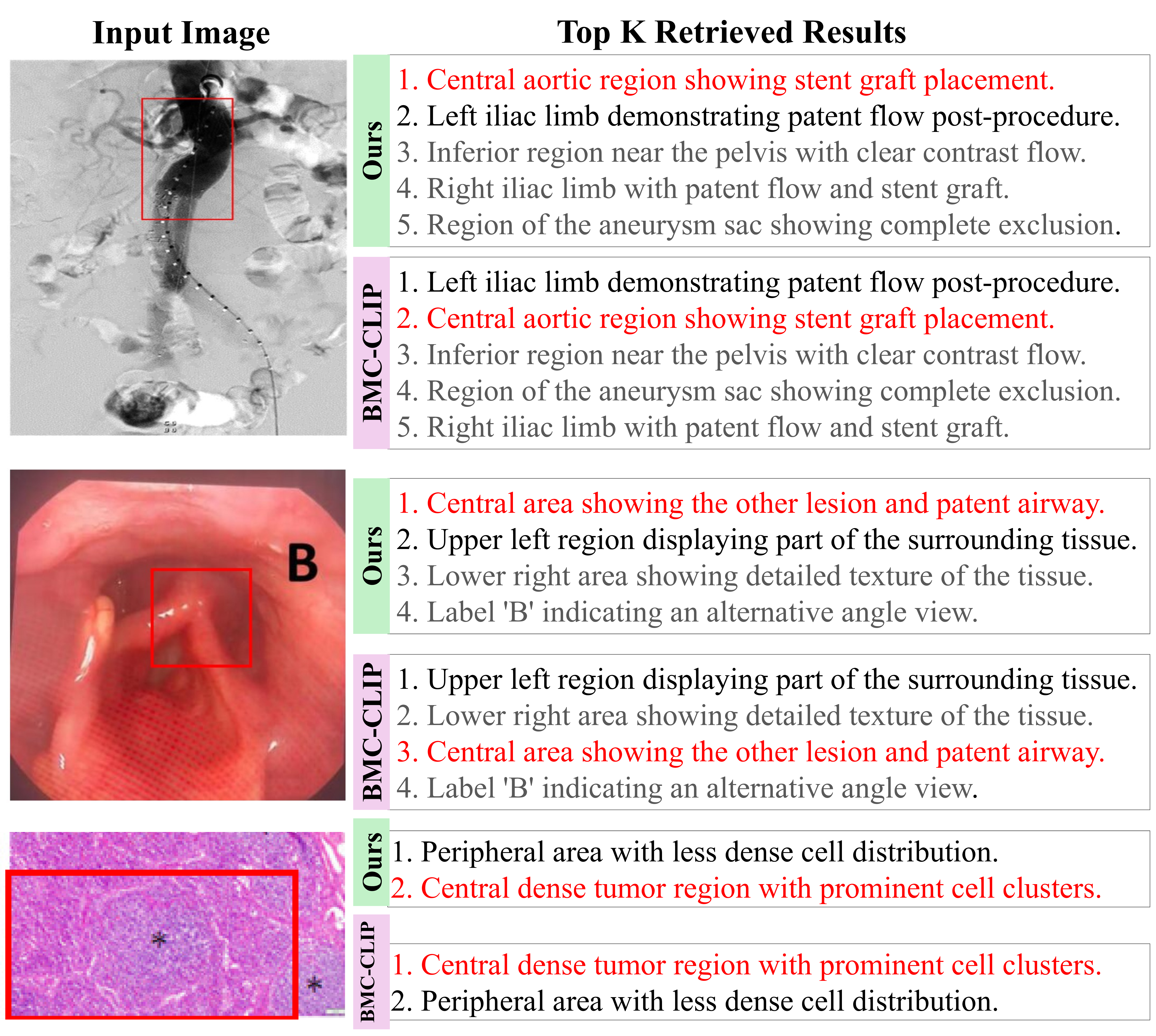
Existing vision-language data generation pipelines face a fundamental tradeoff between scalability and fine-grained supervision, as shown in Tab. 1. Natural computer vision methods [12,30,33,34] achieve fine-grained region-level Table 1. Comparison of vision-language data generation pipelines. Our Panel2Patch produces multi-panel, single-panel, and region-level correspondences, covering all granularities with automatic hierarchical captions and no additional annotation cost. alignment through human annotations, pretrained detectors, or specialized captioners, but such annotation-intensive pipelines incur high computational costs and poorly generalize to biomedical domains due to domain gaps and imaging diversity. Conversely, scalable approaches that mine from biomedical scientific literature sacrifice granularity. They either treat multi-panel figures as single coarse instances [16,44] or decompose them into panels [2] while still retaining figure-level captions that reference multiple elements, thus resulting in weak and coarse image-text alignment and missing the fine-grained region-level correspondences. Fundamentally, none of these methods produce hierarchical supervision spanning all three granularities, i.e., multi-panel figures, individual panels, and finegrained regions, with corresponding hierarchical captions that capture both global context and localized semantics. This absence of multi-level, semantically grounded supervision limits current models’ ability to learn fine-grained visual-textual correspondences in a scalable manner, hindering precise biomedical multimodal understanding.
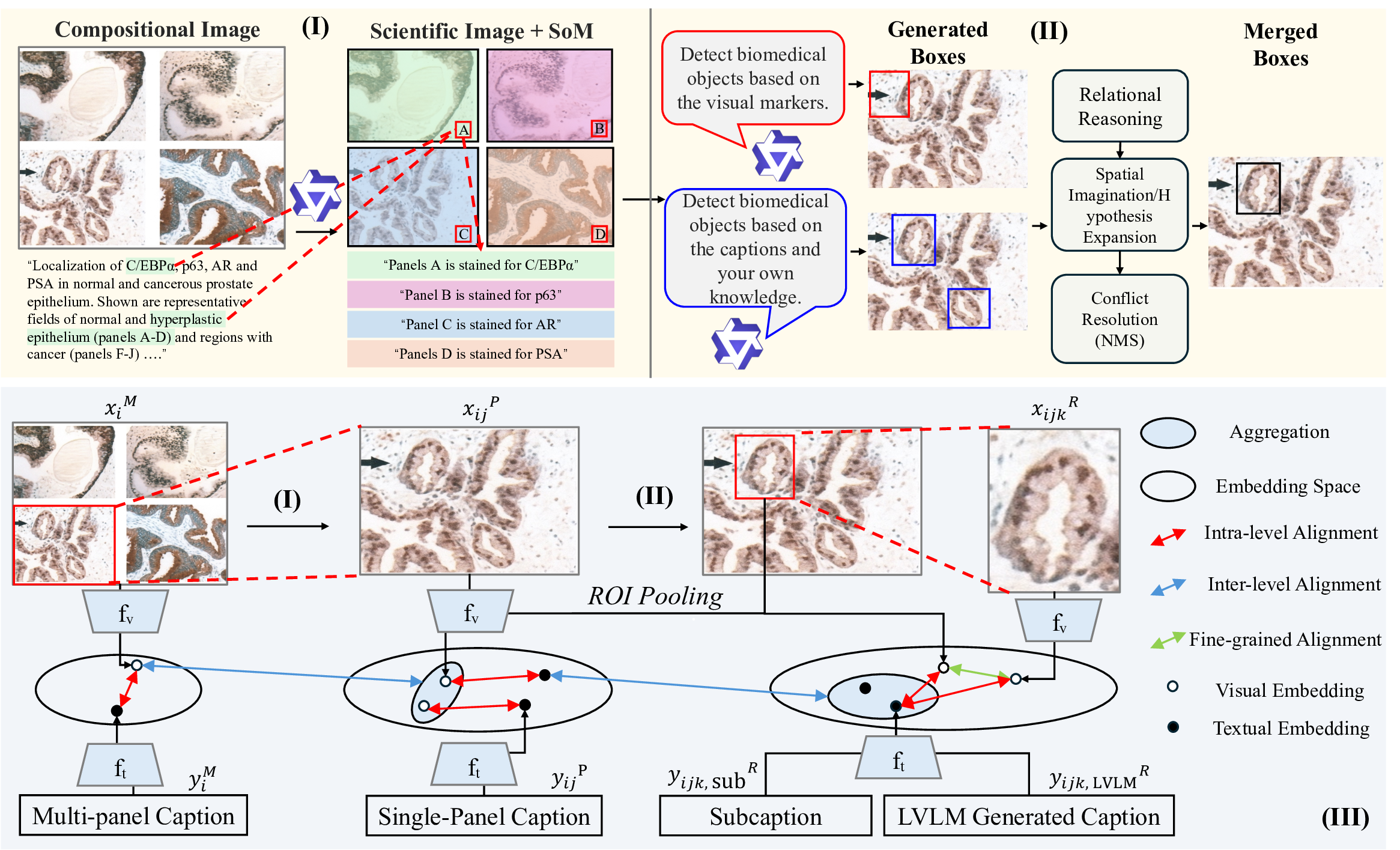
To address these challenges, we propose a comprehensive approach that breaks the scalability-granularity tradeoff through two synergistic contributions. First, we introduce Panel2Patch, a data generation pipeline that exploits a key insight: scientific figures already contain hierarchical visual structure and explicit localization cues that can be automatically extracted. Rather than requiring expensive annotations or specialized detectors, Panel2Patch leverages the inherent pedagogical design of scientific literature, i.e., multi-panel layouts, panel identifiers, and embedded visual markers (arrows, bounding boxes, zoom-in crops), to automatically mine supervision at three granularities: figure, panel, and patch/region. Using off-the-shelf vision-language models with set-of-marks (SoM) prompting, it parses panel structures, decomposes captions, and localizes patches by detecting the visual markers authors use to highlight regions of interest. This yields hierarchical image-text pairs from global figure context to localized patch descriptions, achieving multi-level, fine-grained supervision at scale with minimal cost, as shown in Tab. 1. Panel2Patch establishes a methodology for mining hierar-chical supervision from any domain where visual documentation follows pedagogical conventions, offering a scalable alternative to manual annotation pipelines for biomedicine and other scientific fields to build their foundation models.
We also design a hierarchical zoom-in pretraining framework that uses these multi-granular pairs to train a single CLIP-style encoder whose panel-level embeddings serve as the primary representation for biomedical downstream tasks. The encoder is optimized jointly with auxiliary figure-and patch-level objectives and inter-level message passing, so that panel embeddings are explicitly refined by global figure context and fine-grained regional evidence, through top-down context propagation from figures to panels with global guidance and bottom-up evidence aggregation from patches to panels with localized cues. This hierarchical learning strategy produces a unified encoder where the figure and patch levels serve as contextual scaffolding that enhances the panel-level repres
This content is AI-processed based on open access ArXiv data.